Introduction
Transmissible Spongiform Encephalopathies are rare neurodegenerative brain disorders in both humans (e.g. Creutzfeldt-Jakob disease and Kuru) and animals (Scrapie in sheep, Bovine Spongiform Encephalopathy in cows and Chronic Wasting Disease in deer and elk), characterized by a long incubation period after initial infection. Once symptoms become apparent in humans, the disease progresses inevitably to death within weeks or months, and, to date, no treatment or early preclinical diagnostics are available.
The nature of the infectious agent causing these disorders remains unexplained. The most advertised, but not proven to date, prion protein-only theory simply states that the agent is nothing more than a misfolded host glycoprotein called prion protein1. How this single host protein “encodes” the numerous agent strains that have distinct clinical and pathological features remains to be demonstrated by prion scientists. The infectivity of recombinant prion protein misfolded in a test tube in a mixture with RNA and lipid and later injected into the animal brains was demonstrated a few years ago but was never reproduced independently in a laboratory free of contamination2. The statement that cell free replication of TSE infectivity in a test tube excludes the possibility of the agent being a virus3 ignores the well known fact of human poliovirus replication in a cell free system4.
On the other hand, virino and virus theories claim that host-independent nucleic acid is the genome of the infectious agent5. Virus theory states that the agent is a virus that has not been discovered yet6, while virino theory postulates that the agent is a chimera composed of a host-independent nucleic acid (the genome of the agent) and a host protein, probably the prion protein that protects the genome7. Obviously, nucleic acid-containing theories explain the existence of many agent strains since nucleic acid sequences are the only molecules known to-date that encode phenotypes of all living organisms including microbes, with the smallest among them being the nucleic acids of viroids and satellite RNAs of plant viruses (only few hundred non protein coding nucleotides)8. Despite decades of research efforts, no TSE-specific nucleic acid sequences have been found yet9, leading to the popular conclusion among many scientists that no such nucleic acid exists.
While many different approaches were undertaken to hunt for the elusive viral or subviral nucleic acid, surprisingly, the simplest and easiest of them was not employed. J2 monoclonal antibody recognizes double-stranded RNA (dsRNA) provided that the length of the helix is ≥ 40 bp10. Importantly, dsRNA-recognition is independent of the sequence and nucleotide composition of the antigen. All naturally occurring dsRNA investigated up to now (40–50 species) as well as poly(I)•poly(C) and poly(A)•poly(U) have been recognized by J211. In a systematic study of different viruses, J2 detected dsRNA in cells infected with positive-strand RNA viruses, double-stranded RNA viruses and DNA viruses, but not negative-strand RNA viruses12. This shows that most viruses induce synthesis of long double stranded RNA (dsRNA) during their replication in cells that can be detected by J2. Therefore, the presence of long dsRNA would be an indication of viral infection in cells. J2 antibody has not been used for testing of scrapie infected tissues and the first attempt is made in the present work.
Methods/results
Immunofluorescence
JFH1 Huh7 cells (human hepatoma cells harboring hepatitis C replicon) and Huh7.5 cells (human hepatoma cells free of replicon) were used as a positive (JFH1 Huh7) and negative (Huh7.5) control for dsRNA in immunofluorescence detection experiments (Figure 1a) when probing PK1 cells (a clone of mouse neuroblastoma N2A cells) and RML (Rocky Mountain Laboratory strain of mouse scrapie) infected PK1 (RML/PK1) cells with J2 antibody (mouse monoclonal from Englsih and Scientific Consulting) (Figure 1b). A secondary anti-mouse antibody labeled with Alexa 488 fluorophore is used to visualize J2 binding sites. dsRNA was detected in the cytoplasm of both PK1 and RML/PK1 cells (Figure 1b). The signal was abolished after RNase A (Invitrogen) treatment at 50 µg/ml in 50mM NaCl as it is shown for PK1 cells in Figure 1c.
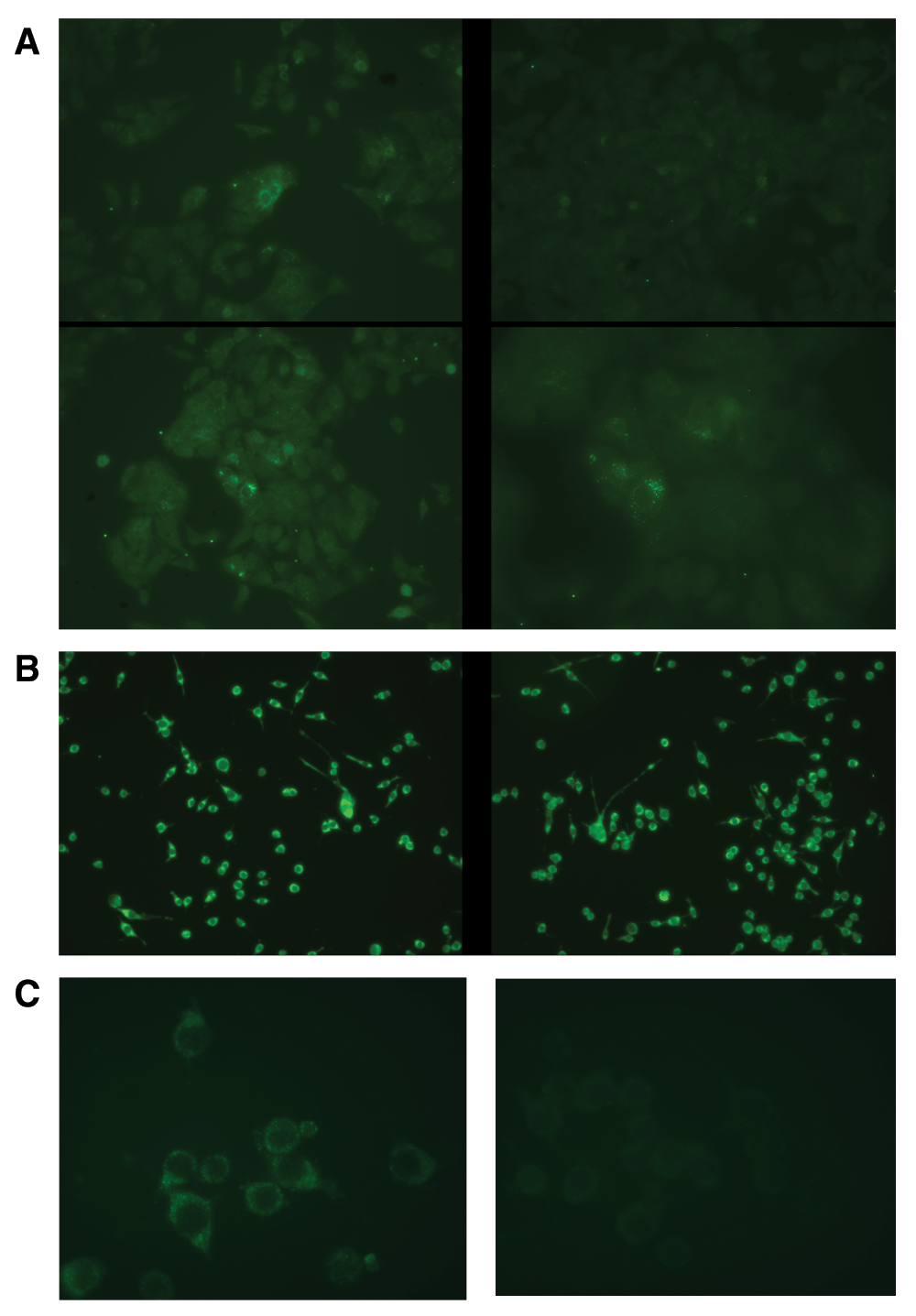
Figure 1. dsRNA is detected with J2 antibody via indirect immunofluorescence.
A secondary anti-mouse antibody labeled with Alexa 488 fluorophore is used to visualize J2 binding sites. (a). In contrast to the absence of green signal in Huh7.5 cells, JFH1 Huh7 cells show punctuate staining in some of them which harbor hepatitis C replicon. (b). dsRNA is detected in both scrapie free PK1 cells and RML scrapie infected RML/PK1 cells. (c). dsRNA disappears from PK1 cells after treatment with RNase A at low salt conditions where it destroys both single stranded and double stranded RNA.
Immunoblotting
J2 antibody was also used for immunoblotting of dsRNA, as described previously13. Crude RNA extracts from JFH1 Huh7, Huh7.5, PK1 and PK1/RML cells were size separated using non-denaturing TBE-polyacrylamide gel, transferred to positively charged Nylon membrane and immunoblotted with J2. Secondary anti-mouse antibody linked to HRP (horseradish peroxidase) and a substrate for it was used for visualization of the blots. Results showed the presence of replication intermediate-RI (upper part of the gel slot, black arrow) and replicative form-RF (seen as a strong band bellow RI, dark blue arrow) in JFH1 Huh7 that were both absent in Huh7.5 (Figure 2). In PK1 and RML/PK1 in addition to dsRNA in the upper part of the gel slots several bands were seen including a duplet with a molecular weight much lower than that of RF of HCV replicon (Figure 2).
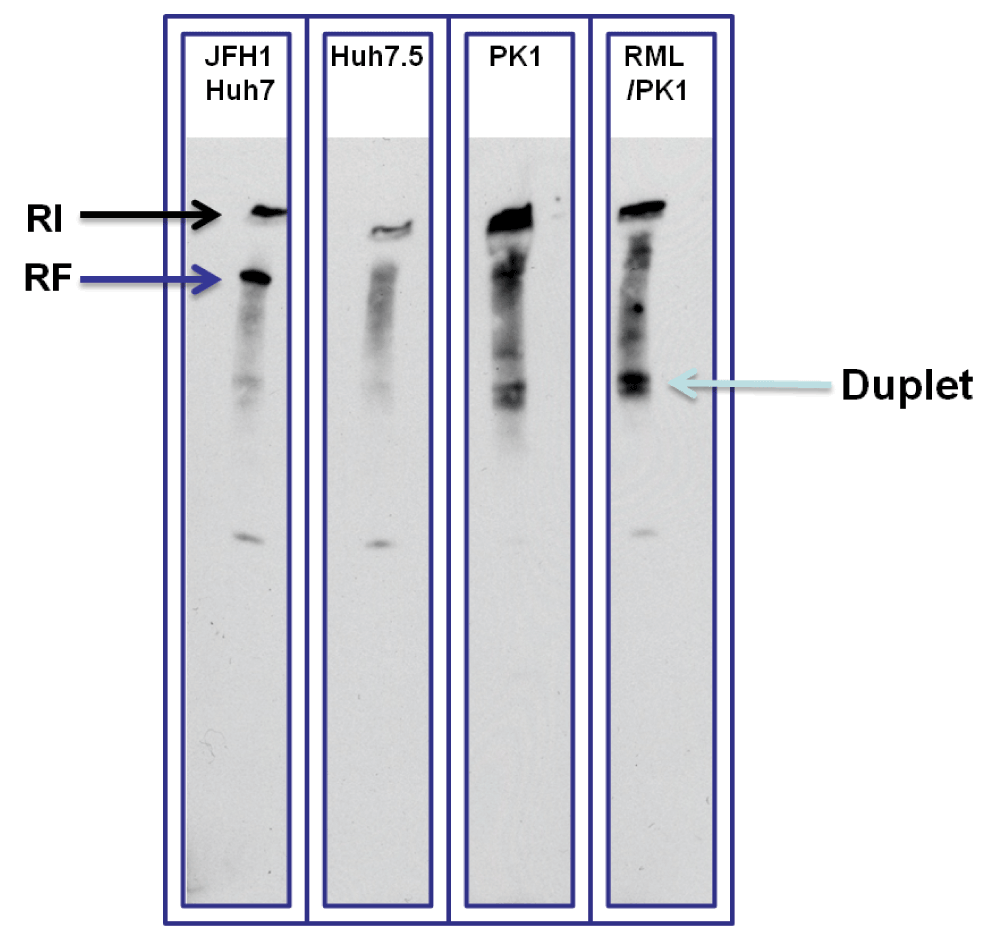
Figure 2. Huh7.5 cells were used as a negative control and JFH1 Huh7 cells as a positive control for dsRNA.
Two bands of dsRNA were detected in JFH1 Huh7 (RI and RF shown by black and dark blue arrows). In PK1 and PK1/RML cells several bands of dsRNA are detected including top ones in the gel slots corresponding to long dsRNA that did not enter the gel. A much lower duplet band could be seen in both specimens. In addition there are several bands in between the duplet and the upper most band.
Immunohistochemistry (IHC)
J2 antibody was recently used for successful detection of viral dsRNA in formalin-fixed paraffin-embedded tissues14. Here an attempt was made to detect dsRNA in 22L scrapie infected mouse brains fixed in Carnoy’s solution and embedded in paraffin. Proteinase K treatment was used as described15, followed by inhibition in glycine (2mg/ml in nanopure water) and short post-fixation in formalin to expose dsRNA for detection. IHC detection of dsRNA in brains using J2 was done with a secondary system described in ref.6. Uninfected C57Bl/6 mice brains were used as control and brains of terminally sick C57Bl/6 mice infected with 22L strain of mouse scrapie were used for the experiment. As a result, dsRNA was detected in scrapie-infected brain predominantly in the cytoplasm of large neurons in the cortex (Figure 3a) and brainstem (Figure 3b). Nuclear staining was also detected in some neurons of the infected brain. In uninfected brain, nuclear staining of some Purkinje cells was detected in the cerebellum (Figure 3c). Otherwise the staining in the control brain was largely absent.
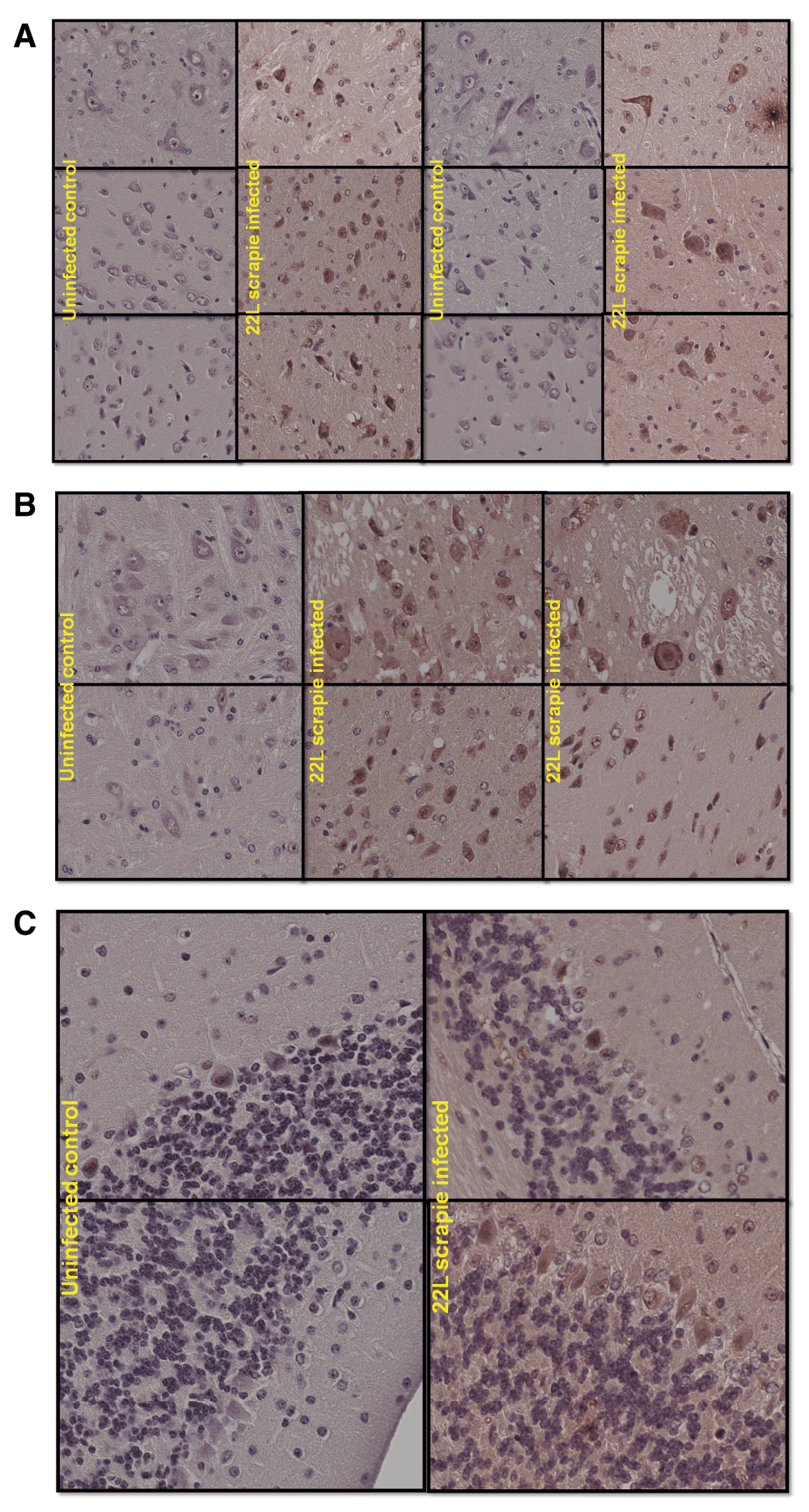
Figure 3. IHC detection of dsRNA in brains using J2 was done with a secondary system described in ref.5.
Uninfected C57Bl/6 mice brains were used as control and brains of terminally sick mice infected with 22L strain of mouse scrapie were used for the experiment. (a). Mostly cytoplasmic and some nuclear staining can be seen in cortical neurons of infected brain. While staining is absent in uninfected control brain. (b). Similar staining pattern is observed in brainstem especially in large neurons. And again staining is absent in parallel uninfected control sections. (c). Some nuclei of Purkinje cells are stained in the cerebellum of both uninfected control and 22L infected brain.
Discussion
Data presented here shows dsRNA is detectable by J2 antibody using immunofluorescence in scrapie susceptible and scrapie infected tissue culture cells. In contrast, it seems that only scrapie infected brain has dsRNA in the cytoplasm of some neurons. Immunoblotting shows long as well as short dsRNA bands in scrapie susceptible and scrapie infected tissue culture cells (Figure 2). Long dsRNA is not present in uninfected mammalian cells and can only be present as a result of viral infection. Therefore long dsRNA presence in scrapie susceptible as well as scrapie infected cells is a strong indication of viral infection of these cells. Shorter dsRNA bands also detected in these cells might point to the presence of subviral nucleic acids16. These data provide an experimental basis for speculation that scrapie agent could be a satellite nucleic acid of a silent and persistent virus that infects susceptible host cells. A virusoid (satellite RNA) in order to replicate would need a helper virus to be in every species and cell that is susceptible to infection. Gajdusek proposed such scenario four decades ago: “These viruses could be associated or satellite viruses which serve to activate or are themselves activated by some helper virus latent in the susceptible host”17.
In analogy to plant satellite RNAs18, scrapie agents’ nucleic acid can function via RNAi to silence host neuronal survival genes (e.g. bcl-2 anti-apoptotic group genes) and cause lethal disease due to its homology with host gene sequences.
Conclusion
For the first time experimental evidence is provided for the presence of long dsRNAs in scrapie infected cells and tissues. This is the strongest argument presented so far for the existence of a virus in scrapie infected cells and tissues. These molecules deserve sequencing and characterization of their relationship to scrapie agent and disease.
Competing interests
No competing interests were disclosed.
Grant information
The author(s) declared that no grants were involved in supporting this work.
Aknowledgments
I thank Dr. Corinne Lasmezas for supervising and supporting my work at Scripps Florida. I am grateful to Dr. Timothy Tellinghuisen, from whom at his lab meeting presentation at Scripps Florida I learnt about the existence of J2 antibody and who sincerely provided human hepatoma cells with and without HCV replicon. I greatly appreciate Franco Sferrazza’s expert help in setting up the immunoblotting experiment.
Faculty Opinions recommendedReferences
- 1.
Prusiner SB:
Novel proteinaceous infectious particles cause scrapie.
Science.
1982; 216(4542): 136–44. PubMed Abstract
| Publisher Full Text
- 2.
Karapetyan Y:
Is Recombinant Prion Protein by Itself Infectious? Reference Source
- 3.
Klingeborn M, Race B, Meade-White KD, et al.:
Lower specific infectivity of protease-resistant prion protein generated in cell-free reactions.
Proc Natl Acad Sci USA.
2011; 108(48): E1244–53. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 4.
Karapetyan YE:
Viruses do replicate in cell-free systems.
Proc Natl Acad Sci USA.
2012; 109(8): E461. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 5.
Dickinson AG, Outram GW:
Genetic aspects of unconventional virus infections: the basis of the virino hypothesis.
Ciba Found Symp.
1988; 135: 63–83. PubMed Abstract
- 6.
Manuelidis L:
A 25 nm virion is the likely cause of transmissible spongiform encephalopathies.
J Cell Biochem.
2007; 100(4): 897–915. PubMed Abstract
| Publisher Full Text
- 7.
Kimberlin RH:
Scrapie agent: prions or virinos?
Nature.
1982; 297(5862): 107–8. PubMed Abstract
| Publisher Full Text
- 8.
Elena SF, Dopazo J, de la Peña M, et al.:
Phylogenetic analysis of viroid and viroid-like satellite RNAs from plants: a reassessment.
J Mol Evol.
2001; 53(2): 155–9. PubMed Abstract
| Publisher Full Text
- 9.
Simoneau S, Ruchoux MM, Vignier N, et al.:
Small critical RNAs in the scrapie agent.
Nature Precedings.
2009. Reference Source
- 10.
Schönborn J, Oberstrass J, Breyel E, et al.:
Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts.
Nucleic Acids Res.
1991; 19(11): 2993–3000. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 11.
http://www.engscicons.de/monoclonal2005_ger/J2_desc2005.htm.
- 12.
Targett-Adams P, Boulant S, McLauchlan J:
Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication.
J Virol.
2008; 82(5): 2182–95. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 13.
Veliceasa D, Enünlü N, Kós PB, et al.:
Searching for a new putative cryptic virus in Pinus sylvestris L.
Virus Genes.
2006; 32(2): 177–86. PubMed Abstract
| Publisher Full Text
- 14.
Richardson SJ, Willcox A, Hilton DA, et al.:
Use of antisera directed against dsRNA to detect viral infections in formalin-fixed paraffin-embedded tissue.
J Clin Virol.
2010; 49(3): 180–5. PubMed Abstract
| Publisher Full Text
- 15.
Karapetyan YE, Saá P, Mahal SP, et al.:
Prion strain discrimination based on rapid in vivo amplification and analysis by the cell panel assay.
PLoS One.
2009; 4(5): e5730. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 16.
Owens RA, Diener TO:
RNA intermediates in potato spindle tuber viroid replication.
Proc Natl Acad Sci USA.
1982; 79(1): 113–7. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 17.
Gajdusek DC:
Spongiform virus encephalopathies.
J Clin Pathol Suppl (R Coll Pathol).
1972; 6: 78–83. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 18.
Smith NA, Eamens AL, Wang MB:
Viral small interfering RNAs target host genes to mediate disease symptoms in plants.
PLoS Pathog.
2011; 7(5): e1002022. PubMed Abstract
| Publisher Full Text
| Free Full Text



In testing a new tool for methods review, I detected the following research resources in your paper which did not contain the RRID (Research Resource IDentifier). As discussed in the instruction ... Continue reading Dear Authors,
In testing a new tool for methods review, I detected the following research resources in your paper which did not contain the RRID (Research Resource IDentifier). As discussed in the instruction to authors, please consider adding these to the final version of your paper.
RRIDs are universal and persistent product codes and are used for antibodies, software tools and model organisms, see instructions to authors:
http://f1000research.com/for-authors/article-guidelines/research-notes
Please verify the accuracy of the RRIDs below:
Needs attention, no match based on information provided:
J2 antibody (mouse monoclonal from Englsih and Scientific Consulting)
please search: scicrunch.org/resources/Any/search?q=J2
A secondary anti-mouse antibody labeled with Alexa 488 fluorophore
please search: scicrunch.org/resources/Any/search?q=Alexa
Uninfected C57Bl/6 mice
please search: scicrunch.org/resources/Any/search?q=C57Bl/6
Please consider adding each research resource highlighted above against the identifier and add the syntax visible in the "cite this" button to the final version of the paper. Note, searching with the catalog number is often the most effective method of quickly finding the RRID.
Kind regards,
Anita
In testing a new tool for methods review, I detected the following research resources in your paper which did not contain the RRID (Research Resource IDentifier). As discussed in the instruction to authors, please consider adding these to the final version of your paper.
RRIDs are universal and persistent product codes and are used for antibodies, software tools and model organisms, see instructions to authors:
http://f1000research.com/for-authors/article-guidelines/research-notes
Please verify the accuracy of the RRIDs below:
Needs attention, no match based on information provided:
J2 antibody (mouse monoclonal from Englsih and Scientific Consulting)
please search: scicrunch.org/resources/Any/search?q=J2
A secondary anti-mouse antibody labeled with Alexa 488 fluorophore
please search: scicrunch.org/resources/Any/search?q=Alexa
Uninfected C57Bl/6 mice
please search: scicrunch.org/resources/Any/search?q=C57Bl/6
Please consider adding each research resource highlighted above against the identifier and add the syntax visible in the "cite this" button to the final version of the paper. Note, searching with the catalog number is often the most effective method of quickly finding the RRID.
Kind regards,
Anita