Keywords
ATP synthase a-subunit, alkaliphile, Lonar Lake India, 16S rRNA, Stenotrophomonas sp. DL18
ATP synthase a-subunit, alkaliphile, Lonar Lake India, 16S rRNA, Stenotrophomonas sp. DL18
In response to the referee comments, changes were
made including modifications in the title, abstract and the discussion along
with the deletion of supplementary figures showing homology modeling of a
subunit, growth studies in alkaline conditions i.e. pH 7.0 to pH 12.0 and the scanning
electron microscopy study. The aim of this updated article is to report on
alkaliphile specific motifs in the a-subunit
of ATP synthase from the isolated alkaliphile, Stenotrophomonas sp. DL18.
Scope of the updated brief report
Lonar Lake in India is a well known source of alkaliphiles. This is the first report from Lake Lonar that is based on alkaliphile specific motifs in the a-subunit of ATP synthase from the isolated alkaliphile, Stenotrophomonas sp. DL18. Although the present study does not have X ray crystallography data of purified ATP synthase and mutational studies, this brief report may initiate investigations of other facultative alkaliphiles research from Lonar Lake
To read any peer review reports and author responses for this article, follow the "read" links in the Open Peer Review table.
In response to the referee comments, changes were made including modifications in the title, abstract and the discussion along with the deletion of supplementary figures showing homology modeling of a subunit, growth studies in alkaline conditions i.e. pH 7.0 to pH 12.0 and the scanning electron microscopy study. The aim of this updated article is to report on alkaliphile specific motifs in the a-subunit of ATP synthase from the isolated alkaliphile, Stenotrophomonas sp. DL18.
Scope of the brief report
Lonar Lake in India is a well known source of alkaliphiles. This is the first report from Lake Lonar that is based on alkaliphile specific motifs in the a-subunit of ATP synthase from the isolated alkaliphile, Stenotrophomonas sp. DL18. Although the present study does not have X ray crystallography data of purified ATP synthase and mutational studies, this brief report may initiate investigations of other facultative alkaliphiles research from Lonar Lake.
ATP is molecular currency for a living cell, which is not only growing and dividing but also continuously responding to external environmental stimuli. To survive in extreme conditions, microorganisms devise specific adaptive mechanisms. Along with other transporter proteins, ATP synthase is widely considered one of the key molecules for adaptation at alkaline conditions. Hydrolysis of nucleoside triphosphates, specifically ATP, provides the chemical energy to drive a wide variety of cellular reactions. ATP synthesis is central to ATP production during oxidative phosphorylation. These are energy-coupling factors and hence called F1Fo-ATP synthases. The Fo integral membrane protein complex provides a transmembrane pore for protons, whereas the peripheral protein F1 is involved in catalysis1. F1 consists of five subunits, α3β3γ1δ1ε1, with a ring of α- and β-subunits alternating around a single γ-subunit2. Fo is a membrane embedded domain with subunits ab2c10–15. Out of these subunits, a-subunit is a stator and c-ring is a rotor ring through which ions (H+ or Na+) are translocated3–6. Each c-chain from the ring consists of two α-helices traversing the membrane and the polar loop extends out of the membrane to interact with the γ-and ε-subunits. A cytoplasmic F1 catalytic domain is connected by a membrane-embedded Fo domain by a central (γε) and peripheral (b2δ) stalk2–6. In general, downhill ion translocation across the membrane through Fo causes rotation of the c-ring, which induces conformational changes in the catalytic β-subunit and results in ATP synthesis. However, in alkaline conditions, the external pH is high i.e. above pH 8.0, which poses a major thermodynamic problem for ATP synthesis. Hence, the cytoplasmic pH needs to be maintained 1.5 to 2.3 pH units below the external environment, which generates an optimal condition for ATP synthesis.
Most studies have focused on the proton translocation channel in the a-subunit of ATP synthase7–9. However, similar experimental evidence from other geographic locations such as highly alkaline soda lakes needs further exploration to understand pH homeostasis in facultative alkaliphiles. This study explores the ATP synthase a-subunit of a facultative alkaliphilic aerobe isolated from Lonar Lake.
In the present study, underwater sediment soil samples were collected 350 meters away from the Kamalaja Devi Temple end of Lonar Lake, Buldhana, Maharashtra, India. The initial screening was performed at pH 9.5. Then isolates were further studied in the range of pH 7 to pH 12. Across the pH gradient, morphologically different pink, orange and white bacterial colonies were observed. The bacterial genomic DNA of optimally grown bacteria at pH 9.5 with orange pigmentation was isolated by the DNAzol method10.
Further polymerase chain reaction (PCR) and sequencing of 16S rRNA was carried out for identification of bacterium11.
The following primers (Integrated DNA Technologies, USA) were used for the amplification of the 1.7 Kbp PCR product of the ATP synthase Fo subunit (consisting of atpB, atpC and atpA genes), based on a reference strain (i.e. S. maltophilia K279a): forward primer Steno atp1F (5’ CCTGGCGGATCCTTAGATCTCCG 3’) and reverse primer Steno atp1R (5’ CAGTGAGGATCCTTAGATCTCCGAGGCCAGCT 3’). Briefly, a PCR reaction mixture of 100 µl was prepared: 10x PCR buffer, 50 mM MgCl2, 10 mM dNTP mix, 100 picomoles each of forward and reverse primers, 5 units of Taq DNA polymerase (Chromous Biotech, India) with Pfu and 200 ng of bacterial genomic DNA template in nuclease free water. The thermal cycling conditions were: initial denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30s, primer annealing at 55°C for 30s and primer extension at 72°C for 3 min. A final extension was carried out at 72°C for 5 min. The amplified PCR product of 1.7 kbp was visualized on 1% agarose gel and the results were documented by Bio-Rad gel documentation system with Quantity One software (Bio-Rad, USA). Further, the DNA sequencing of the ATP Fo subunit of the selected isolate was performed via primer walking. An ATP synthase comparative study was performed by multiple sequence alignment with other strains from different categories i.e. acidophiles, neutrophiles and alkaliphiles, as shown in Figure 1.
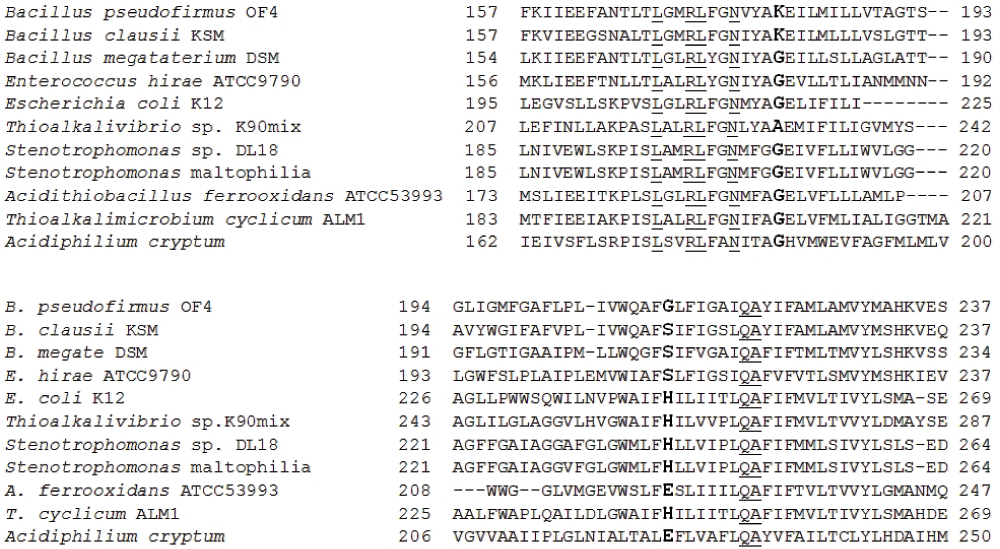
Conserved sequences are underlined and significant variant residues for channel formation are shown in bold. These residues are mainly from TMH-4 and TMH-5. Bacillus megaterium DSM 319 (GenBank Accession number: YP_003600289) grows strictly in neutral pH environments. However, Bacillus clausii KSM K-16 (GenBank Accession number: YP_177349) is a reported alkaliphile.
The orange pigmented bacterium was identified as Stenotrophomonas sp. based on a NCBI BLAST analysis of the 16S rRNA gene sequence and titled Stenotrophomonas sp. DL18 (GenBank Accession number: JN995612). Microbiological studies we performed showed that Stenotrophomonas sp. DL18, which optimally grows at pH 9.5, is an aerobic, facultative alkaliphilic with curved rod morphology.
A BLAST analysis of the ATP synthase a-subunit of the Stenotrophomonas species DL18 suggests maximum identity with Stenotrophomonas species SKA14 (GenBank Accession number: ZP_05136035) at the amino acid level (259 amino acid residues were identical from a total of 266 amino acid residues in the SKA14 species).
Table 1 lists the bacterial species used in the comparative studies. The amino acid residue arginine, which was found to be conserved in almost all bacterial species in the a-subunit, was observed at position 200 (Arg200) in DL18. Moreover, other amino acid residues that were conserved in most of the bacteria include Leu207, Arg210, Leu211, Gly213, Asn214, Gly218, Gln252, Ala253 and Phe255 (E. coli numbering system for ATP synthase a-subunit) in the trans-membrane helix-4 (aTMH-4), aTMH-5 and the corresponding amino acids were also found in the DL18 alignment (Figure 1 and Figure 2).
| Species | Accession number |
|---|---|
| Stenotrophomonas sp. DL18 | AET99165 |
| Stenotrophomonas sp. SKA14 | ZP_05136035 |
| Bacillus pseudofirmus OF4 | YP_003426326 |
| Bacillus clausii KSM-K16 | YP_177349 |
| Enterococcus hirae ATCC 9790 | YP_006487510 |
| Bacillus megaterium DSM 319 | YP_003600289 |
| Stenotrophomonas sp. K279a | YP_001973793 |
| Theoalkalivibrio sp. K90mix | YP_003461818 |
| Escherichia coli K12 DH10B | YP_001732559 |
| Acidithiobacillus ferrooxidans ATCC 53993 | YP_002221206 |
| Acidiphilium cryptum JF-5 | YP_001233541 |
| Thioalkalimicrobium cyclicum ALM1 | YP_004537849 |
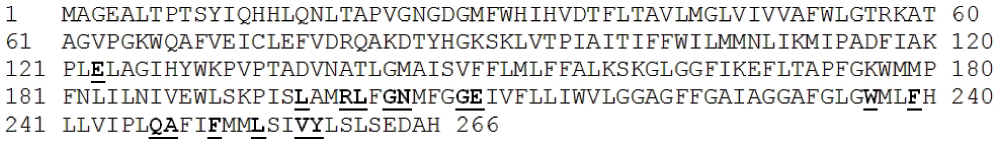
The most conserved amino acid residues are shown as bold and underlined.
It was observed that the most conserved arginine residue of the a-subunit (Arg200 of Stenotrophomonas sp. DL18) was aligned with the expected position of the facultative alkaliphile Bacillus pseudofirmus OF4 (i.e. Arg172). This positively charged Arg200 plays an elemental role in the function of the Fo rotor3. Lys180 in B. pseudofirmus OF4 was replaced by Gly208 in the DL18 strain. In addition, glycine and alanine residues were observed at the same position in other alkaliphiles including Gly206in Thioalkalimicrobium cyclicum ALM1, and Ala230 in Theoalkalivibrio sp. K90mix, as shown in Figure 1. On the other hand, glycine was also at the same corresponding position in alignment for the acidophiles, Acidithiobacillus ferrooxidans ATCC 53993 and Acidiphilium cryptum JF-5 (Figure 1). As reported by Ivey DM et al., Lys180and corresponding amino acids were located in aTMH-4 in Bacillus pseudofirmus OF4 and other alkaliphiles12,13. In the DL18 strain, a histidine residue, which is conserved in other reference species of the same genus (e.g. Stenotrophomonas species K279a, Stenotrophomonas sp. SKA14 and other alkaliphiles including T. cyclicum ALM1 (His244), and Theoalkalivibrio sp. K90mix (His262), was present at position 240 (His240) (Figure 1).
It has been proposed that Gly120 and Lys180forms a channel for the proton uptake pathway of the a-subunit through which protons pass onto the neighboring c-subunit in B. pseudofirmus OF47. However, one study has reported that His245 along with the Gly218 and Glu219 positioning plays a critical role in Fo ion translocations in E. coli8, and in the present study on Stenotrophomonas sp. DL18, His240, Gly208 and Glu209were observed at the same corresponding positions as shown in Figure 1. McMillan14 et al demonstrated the importance of residues with a basic side chain along with its pKa value in ATP synthesis in alkaline environments. However, that mutation study was carried out in the Bacillus sp. TA2.A1, specifically involving the 180th residue in the a-subunit. In that amino acid substitution study, amino acid residue Lys180 in the a-subunit was mutated to Gly180, His180and Arg180. These mutations indicated the ATP synthesis at neutral, neutral to alkaline and only in alkaline conditions (>pH 8.5), respectively, i.e. residue with a strong base, such as lysine or arginine, was ideally appropriate to function at alkaline pH. This was particularly marked for histidine, which has a pKa in a neutral range, showing significant ATP synthesis activity at pH 9.0.
E. coli K12 DH10B considered a neutrophile, can adapt to slightly alkaline conditions up to pH 8.0 and this may be due to the presence of His245. In addition, exchange mutations at aE219-H and aH245-E showed similar ATP synthase activity in E. coli. Moreover, the aG218-K substitution effect was suppressed by aH245-G mutation in E. coli. Hence, these studies in E. coli signified that the positions Gly218and His245 along with Glu219had a critical interaction with the Fo subunit function. In addition, equivalent amino acid residue studies were also reported from the facultative alkaliphile B. pseudofirmus OF4 a-subunit7–9.
From alignment, Glu209 of the DL18 strain was conserved in alkaliphiles, neutrophiles and some acidophiles except His186in Acidiphilium cryptum (Figure 1). In addition, the position of Gly212 in B. pseudofirmus OF4 corresponds to Ser212 in Enterococcus hirae ATCC 9790 (neutrophile), His245 in E. coli and His240 in the Stenotrophomonas sp. DL18 while glutamate in acidophiles is positioned at Glu222 in A. ferrooxidans and Glu225 in A. cryptum as shown in Figure 1. This showed that channel formation may involve a glycine residue along with other residues, specifically those with acidic, basic and neutral side chains, which play vital roles in ATP synthesis in acidophile, alkaliphiles and neutrophiles. Hence, these residues are found to be critical in channel formation7–9. Similarly, in Stenotrophomonas sp. DL18, amino acid residues. Gly208 and His240may form the proton translocation channel. Thus, the basic side chain residue His240 and other amino acid residues may responsible for growth of the Stenotrophomonas sp. DL18 at high alkaline pH.
Devendra Lingojwar contributed to the conception and design of the project, acquired, analyzed and interpreted the data, review of literature, and prepared the manuscript. Ravikant Jadhav helped prepare the manuscript, contributed to data collection and conducted Bioinformatics analysis. Kachru Gawai provided overall guidance for the project, helped prepare the manuscript and revised it critically for intellectual content. All authors approved the final version of the article.
Specific facilities related to molecular biology were kindly provided by ATG LAB. No specific grant was available for this project. The entire project was carried out with the support from the ATG LAB as an intramural project.
Authors are thankful to the Department of Chemistry, University of Pune, India for providing the place of research and infrastructure and ATG LAB for providing laboratory facilities whenever needed. The authors are also thankful to Mrs. Sarita Lingojwar, Admin Head and Laboratory Manager, ATG LAB, for providing kind help and support for laboratory facilities during this project.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (update) 26 Apr 13 |
read | |
|
Version 1 07 Dec 12 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Both the referees have queries and objections as to why crystallographic studies were not reported and why mutation studies were not reported. I ask a question, how many alkaliphiles were reported from alkaline sources without alkaliphile specific sequences? What is mentioned in this updated version of this article is all about reporting an alkaliphile from the source known for decade. But at the molecular level, studies on Lonar Lake alkaliphiles are incompletely understood or rather not reported earlier.
Both the referees have queries and objections as to why crystallographic studies were not reported and why mutation studies were not reported. I ask a question, how many alkaliphiles were reported from alkaline sources without alkaliphile specific sequences? What is mentioned in this updated version of this article is all about reporting an alkaliphile from the source known for decade. But at the molecular level, studies on Lonar Lake alkaliphiles are incompletely understood or rather not reported earlier.