Keywords
Citrus greening disease, cuttings, latent infection, real-time PCR
This article is included in the Agriculture, Food and Nutrition gateway.
Citrus greening disease, cuttings, latent infection, real-time PCR
Citrus greening disease (huanglongbing; HLB) is one of the most devastating diseases for the citrus industry worldwide and is caused by the phloem-limited phytopathogenic bacteria ‘Candidatus Liberibacter’ spp. (Bové, 2006; Jagoueix et al., 1994). In particular, ‘Candidatus Liberibacter asiaticus’ (CLas) has been an important causal agent in many countries including parts of Asia (Bové, 2006). In Japan, the occurrence of citrus greening disease has been limited to areas of the south including the Okinawa prefecture and Oshima Islands, with Tokunoshima Island being the upper limit (Moji Plant Protection Station, 2012). Disease management has been carried out to prevent a further spread of the disease to citrus production areas on the main islands. A combination of PCR techniques for CLas detection and the disposal of infected citrus trees is the most practical approach to control the disease as there is currently no cure. On Kikaijima Island, located among the Amami Islands, a program for eradication of citrus greening disease was conducted in 2005 and had accomplished the eradication of the disease by 2012, resulting in an official declaration of eradication on Kikaijima Island (Moji Plant Protection Station, 2012). Currently, the eradication program is in progress on the larger Island of Tokunoshima, which is located to the south of Kikaijima Island.
Citrus greening disease is not often distinguishable from physiological disorders. Early symptoms of citrus greening disease appear in the leaves and include yellowish, blotchy chlorosis and mottling of leaves (Bové, 2006). On the other hand, these symptoms resemble other disorders including virus infections and nutrient deficiencies (Tian et al., 2014). Also, the disease primarily involves latent infection of CLas. In this asymptomatic condition, CLas is often undetectable with PCR techniques because the pathogen has infected the asymptomatic tree at a very low density (Gottwald et al., 2007). Asymptomatic trees that are overlooked in the fields are an important source of infection through vector-mediated transmission by the Asian citrus psyllid, Diaphorina citri Kuwayama (Hemiptera: Psyllidae) (Pelz-Stelinski et al., 2010). Because current disease management only allows us to identify citrus greening disease when CLas is detected by PCR and real-time PCR, the sensitivity and robustness of PCR detection is indispensable (Arredondo Valdés et al., 2016; Folimonova & Achor, 2010; Fujikawa & Iwanami, 2012). Recent studies have made significant progress in understanding the primary localization of CLas in roots before systemic distribution (Louzada et al., 2016; Park et al., 2018). CLas density in roots is relatively higher than that in leaves (Johnson et al., 2014). This suggests that root samples would be an alternative target for early detection of CLas. On the other hand, one drawback to using crude roots for detecting CLas would be the difficulties in processing DNA-based techniques. In contrast to leaf samples, root samples are not easily available in field surveys because of the hardened tissues in mature citrus trees, and it may be a struggle to dig up root tissues for further assays. In this study, we developed a new approach for early detection of CLas using root samples recovered from cuttings of CLas-infected trees. This approach allowed us to detect CLas from asymptomatic citrus trees that had previously tested negative when using leaves for conventional PCR and real-time PCR.
One-year-old seedlings of rough lemon (Citrus jambhiri Lush.) were prepared in 12 cm-diameter × 24 cm-deep pots. Graft-inoculation of 12 seedlings was carried out with a small amount of bark tissue of CLas-infected flat lemon trees (Citrus depressa Hayata.), which has been maintained in a quarantined greenhouse under a special permission (26Y1214) issued by the Ministry of Agriculture, Forestry and Fisheries (Fujikawa et al., 2013). CLas infection on seven rough lemon trees was confirmed by both the development of foliar symptoms and the presence of CLas by real-time PCR using HLBas/HLBr/HLBp primer–probe set (Li et al., 2006). Five rough lemon trees did not develop any foliar symptoms, and CLas was undetectable from leaves over two years in a monthly survey with both the conventional PCR (Fujikawa et al., 2012) and real-time PCR using HLBas/HLBr/HLBp primer–probe set (Li et al., 2006). These inoculated but asymptomatic rough lemon trees (five pots in all) were used as asymptomatic trees for this study.
Cuttings were prepared from the upper part of young green shoots and branches, which were taken from seven symptomatic and five asymptomatic rough lemon trees. Cuttings of approximately 4 cm in length containing one leaf with no disease symptoms were selected and treated with 1% “Menedael” Fe (II) aq. fertilizer (Menedael Co., Ltd., Osaka, Japan; Cat. No. 242461) and then dipped in “Rooton”, a synthetic auxin α-naphthyl acetamide, (Sumitomo chemical garden products inc., Tokyo, Japan; JAN code 4975292090625) for promotion of rooting. The cuttings that had been treated were planted in a box that contained autoclaved “Kanuma” pumice and kept at approximately 32°C for a month under natural light/dark conditions, covered with a plastic film. Care was taken to keep the Kanuma pumice gently wet but not saturated to ensure optimum rooting. Roots that regenerated a month after cutting were used for CLas detection in the real-time PCR (Li et al., 2006).
For detecting CLas in cuttings with root-sprouts that tested negative in the first PCR test, subsequent cultivation of the cuttings in various soils was carried out in a quarantined greenhouse and monitored for two months. Five cuttings were individually transplanted into the following soils: two commercial soils which were “Kenbyo” nursery soil (Yaenougei Co., Ltd., Nagasaki, Japan) and “Sanyo composted bark” soil (Sanyo Chip Co., Ltd., Yamaguchi, Japan); one natural soil from an area where citrus greening disease has occurred, namely “Kunigami Maaji” clay soil collected from Kunigami-son village (around N26°44'44.8", E128°10'42.0"), Okinawa Prefecture; and two natural soils from areas where citrus greening disease has not occurred, which were Acrisols collected from Isen-cho (around N27°40'25.2", E128°56'16.2"), Tokunoshima Islands, Kagoshima Prefecture and Andosols collected from Tarumizu-city (around N31°28'38.1", E130°43'19.5"), Kagoshima Prefecture. The natural soils were collected in 2016. During soil sampling, the surface litter and soil at the sampling spot was removed using a spade. Using a plow at each sampling site, each natural soil was collected up to a depth of 10 cm and bulked. The precise locations of the sampling sites are not disclosed due to landowner privacy reasons. All soils were prepared with and without autoclaving. The autoclaving process was repeated twice. During the cultivation, 0.1g of “Hyponex” powder fertilizer (HYPONeX Japan Co., Ltd., Tokyo, Japan; JAN code 4977517003052) and one tablet of Aid-ball Ca (Sumitomo chemical garden products Inc., Tokyo, Japan; JAN code 4975292110118) were applied once a month. A single leaf and root were collected from each cutting on a monthly basis to determine the CLas infection status using real-time PCR (Li et al., 2006).
Leaf samples were cleaned and wiped with a kimwipe paper (Kimberly Clark Corp., Irving, TX) impregnated with 70% ethanol. Midribs were excised and chopped into 2–3 mm pieces, providing approximately 0.05 (±0.01 SD) g of the midribs for DNA extraction. For the root samples, a single root was vortexed vigorously to remove soils. Approximately 4 cm of the root segment was cut out from the bottom of the root and then wiped with a kimwipe impregnated with 70% ethanol. All sample materials were individually ground with a mortar and pestle, and then subjected to DNA extraction using the commercial kit DNeasy Plant Mini Kit (QIAGEN, Valencia, CA; Cat. No. 69106) according to the manufacturer’s instructions. DNA was eluted in 200 μL of a given AE buffer. CLas detection in cuttings was performed by TaqMan probe real-time PCR in a QuantStudio 5 real-time PCR system (Thermo Fisher Scientific Inc., Waltham, MA) using the HLBas (5’-TCGAGCGCGTATGCAATACG-3’) / HLBr (5’-GCGTTATCCCGTAGAAAAAGGTAG-3’) / HLBp (6-FAM-AGACGGGTGAGTAACGCG-BHQ-1) primer–probe set (Li et al., 2006). Premix Ex Taq (Probe qPCR) (Takara Bio Inc., Shiga, Japan) was used according to the manufacturer’s protocol. The PCR reaction volume of 20 µL contained 0.4 µL of each primer, 10 µL of Premix Ex Taq, 0.8 µL of HLBp, 0.4 µL of ROX Reference Dye II, 2 µL DNA template, and 6 µL of sterile distilled water. The real-time PCR consisted of pre-denaturation at 95°C for 30 s, and 45 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min. CLas concentration was calculated based on the relationship between quantity and molarity of CLas 16S rDNA (as outlined by Degen et al., 2006), and the standard curve analysis was determined using the amount of template DNA adjusted to the DNA content in 108 cells/μL and a 10-fold dilution series. The CLas density in leaves was calculated from the total amount of DNA eluted from leaves according to the representative standard curve (y = −1.538x + 42.278, R2 = 0.9999). The results of the CLas development were analyzed using Tukey-Kramer multiple comparison test in the R statistical software (v. 3.5.0)
Differences were considered significant when P < 0.01.
Most of the cuttings from asymptomatic trees survived and were well rooted (Figure 1a–c), but the regeneration of cuttings using symptomatic trees was evidently influenced by CLas infection. A total of 224 clones regenerated from 240 prepared cuttings using asymptomatic trees whereas a total of 24 clones were recovered from 45 prepared cuttings using symptomatic trees. One root sample was collected from each cutting and these were used for the primary PCR test. CLas was detected in a total of 115 cuttings in the real-time PCR (Fujiwara, 2021a), which corresponds to a 51.3% detection rate (Table 1), while none of the cuttings showed foliar symptoms of citrus greening. In addition to this test, nine leaf samples from cuttings showing CLas negative in roots were randomly selected and subjected to real-time PCR, the results of which showed no detection of CLas within the leaf samples. In regeneration of cuttings using symptomatic trees, CLas was detected in all root samples, which corresponds to a 100% detection rate (Table 1), as was detected in the symptomatic leaves.
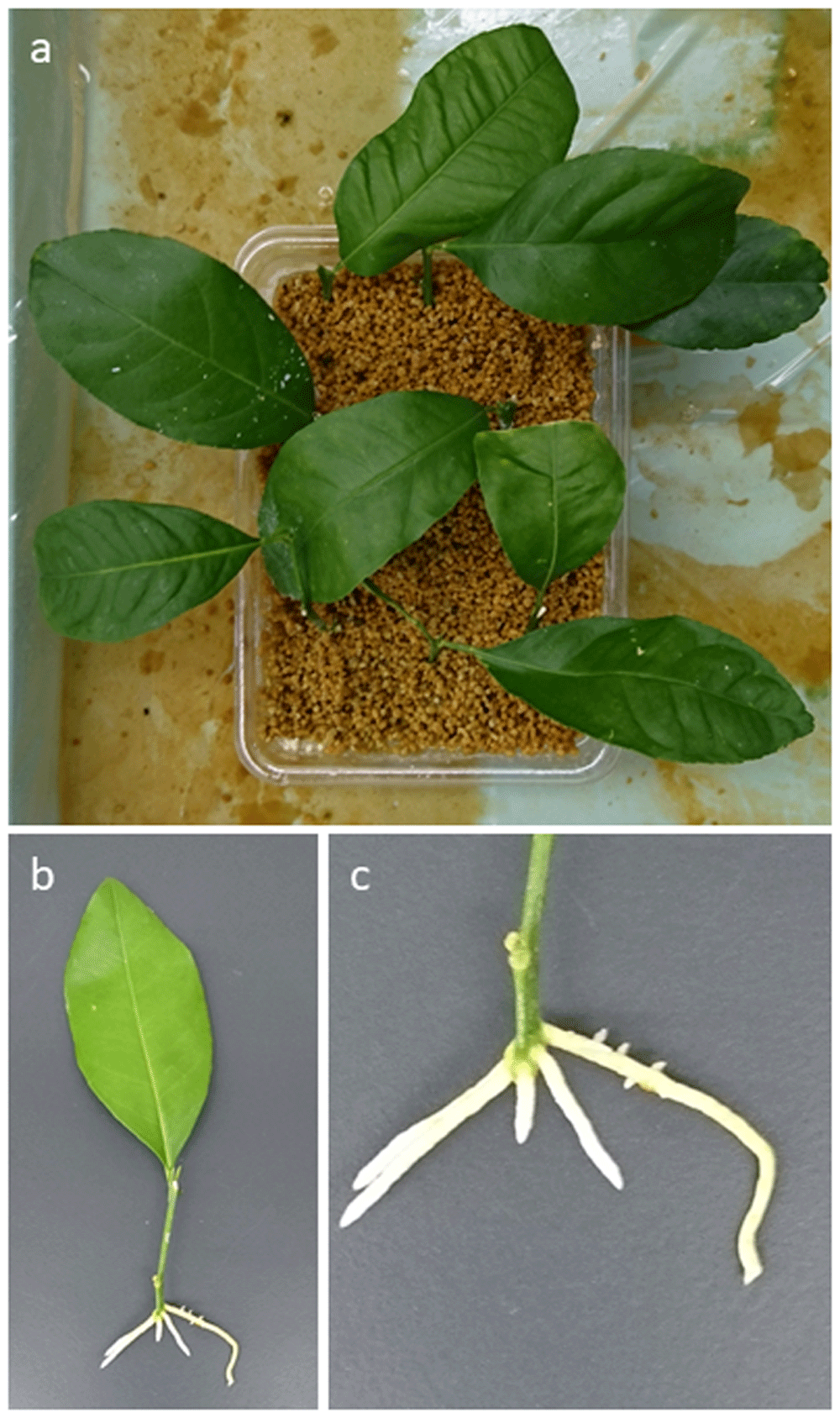
Preparation of cuttings using asymptomatic trees; a, cuttings grown in Kanuma soil a month after preparation; b–c, seedlings recovered from cuttings showing newly developed roots.
| Root samples* | CLas detection | |||
|---|---|---|---|---|
| + | - | |||
| Seedlings recovered from cuttings | Asymptomatic | 224 | 115 (51.3%) | 109 (48.7%) |
| Symptomatic | 24 | 24 (100%) | 0 | |
* Roots re-generated a month after cutting were used to detect CLas by the real-time polymerase chain reaction. Raw data is provided on Figshare (Fujiwara, 2021a).
A subsequent cultivation test was performed (Fujiwara, 2021b) to assess the CLas recovery from latent infection using the cuttings which showed no CLas detection in the primary PCR test (Supplementary Figure 1, Extended data (Fujiwara, 2020)). To examine suitable soil types for CLas development in cultivation, we tested five different types of soils including two commercial soils (“Kenbyo” nursery soil and “Sanyo composted bark” soil) and three natural soils (Acrisols, Andosols, and “Kunigami Maaji” soil). Overall, a cultivation of the cuttings in prepared soils successfully provided CLas detection by real-time PCR from asymptomatic trees (Figure 2). In the nursery soil, CLas was detectable from both leaves and roots a month after transplanting (P < 0.01). The density of CLas in seedlings grown in the “Kenbyo” nursery soil with autoclaving (3.4 × 105 cells/leaf and 2.1 × 105 cells/root) was relatively higher than that in the same soil without autoclaving (3.8 × 103 cells/leaf and 4.9 × 103 cells/root), and the detection was stable throughout the cultivation for two months (Figure 2; Supplementary Figure 2, Extended data (Fujiwara, 2020)). In contrast, although CLas was likely to be detected in both leaves and roots grown in a composted soil (1.7 × 103 cells/leaf and 4.5 × 101 cells/root with autoclaving), the CLas density detected in cultivations in the composted soil without autoclaving fluctuated highly (Figure 2; Supplementary Figure 3, Extended data (Fujiwara, 2020)). Seedlings grown in composted soil without autoclaving showed CLas development a month after cultivation, no CLas was detected in the subsequent cultivation. In the natural soils, which comprised Acrisols, Andosols, and “Kunigami Maaji” soils, it was also the case that CLas detection fluctuated in both the leaves and roots similar to that in the composted bark soil without autoclaving (Figure 2; Supplementary Figure 4–6, Extended data (Fujiwara, 2020)). Autoclaving of the Andosols and “Kunigami Maaji” soil had little effect on the propagation of CLas in the cuttings, showing 2.3 × 103 cells/leaf and 1.1 × 104 cells/root in Andosols without autoclaving; 5.2 × 102 cells/leaf and 9.5 × 102 cells/root in a Kunigami Maaji soil without autoclaving. In Acrisols, CLas development was relatively varied compared with other natural soils and appeared to be affected by the soil conditions with autoclaving (5.4 × 103 cells/leaf and 2.2 × 104 cells/root with autoclaving) (Figure 2; Supplementary Figure 4, Extended data (Fujiwara, 2020)). In short, the soil conditions of a nursery soil treated with autoclaving was suitable for early detections of CLas recovered from latent infection using cuttings.
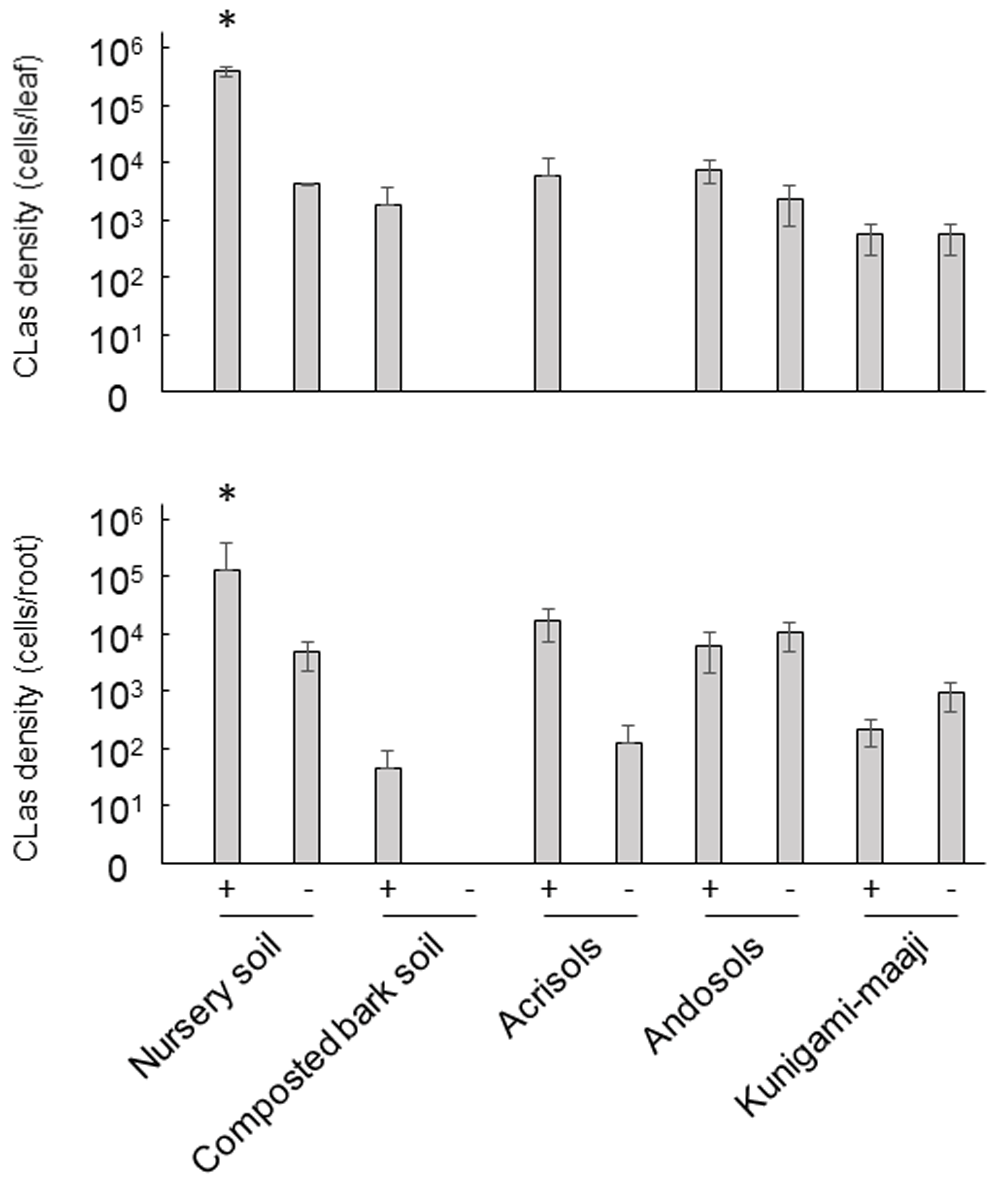
Average of CLas density two months after cultivation was determined by real-time PCR and calculated using CLas density in five individual seedlings shown in Supplemental Figures 2–6 (see Extended data). CLas recovered from latent infection was likely to be detected from leaves and roots using nursery soil with autoclaving. Statistical significance was shown (*P < 0.01, Tukey-Kramer multiple comparison test). +: with autoclaved, -: without autoclaved.
This study showed that while it remains difficult to identify CLas from latent infection in host trees, latently infected CLas can be detectable by real-time PCR using roots regenerated from cuttings and grown in autoclaved nursery soil. The results indicated that detection from cuttings with or without cultivation in autoclaved nursery soil can be practically applicable to detect CLas from latently infected trees. In field tests, it might be possible to detect CLas directly from the root samples of latently infected trees, but in some seasons and locations digging on site and finding new, fine, roots would be extremely difficult. Instead, the CLas detection from cuttings reported here is suitable in any surrounding as long as twigs are available.
Soil chemicals and microorganisms evidently affect the occurrence and distribution of CLas in citrus (Blaustein et al., 2017) as is the case with other soil-borne diseases (Philippot et al., 2013). CLas is likely to accumulate in roots prior to systemic distribution in citrus trees (Johnson et al., 2014) and requires CLas-associated microbiota in the host plant for its own survival (Fujiwara et al., 2018). This suggests that disturbance of CLas-associated microbiota by soil microorganisms may affect CLas development in citrus trees. A subset of CLas-associated microbiota in roots was structured by CLas infection, resulting in distinct microbial and metabiological profiles in roots (Padhi et al., 2019; Zhang et al., 2017). Thus, soil and rhizosphere management may be critical for disease control management. On the other hand, there is a concern that improved root integrity slows CLas growth and delays disease development, making it difficult to detect CLas in roots and other plant tissues. Our study has shown that CLas recovery from latent infection varied with soil type and conditions. While we still need to clarify the interplay between soil functions and CLas behaviors, further accumulation of knowledge on the relationship between root zone and the occurrence of citrus greening diseases is expected to enable more effective monitoring and detection of CLas in roots (Blaustein et al., 2018).
A citrus greening disease eradication program in Japan has been promoted by both the early detection of CLas from leaf samples using PCR methods and the removal of the infected trees. Detection of CLas from latent infection was once carried out by a validation method using grafting over six months (Figure 3a). The disease management has been successful in increasing citrus greening disease-free areas in the Okinawa prefecture and Amami Islands. In addition to the conventional survey, a new approach developed in this study could be provided as part of an eradication program for finding latent infection of CLas and is expected to contribute to time-and cost-efficient validation of CLas infection detection. In practice, the process of a new approach consists of two sequential steps (Figure 3b; Supplementary Figure 7a-b, Extended data (Fujiwara, 2020)). As foliar samples from an asymptomatic tree test negative in the primary PCR test, cuttings should be prepared from the asymptomatic tree. Roots regenerated from the cuttings are then used for CLas detection in the second PCR test (Figure 3b; Supplementary Figure 7a, Extended data (Fujiwara, 2020)). If CLas is detected from any of the cuttings, the asymptomatic tree would be removed from the field. Next, a subsequent cultivation test is conducted as the third step if all cuttings from the asymptomatic tree test negative in the second PCR test (Figure 3b; Supplementary Figure 7b, Extended data (Fujiwara, 2020)). Root samples are collected within a few months after cultivation, and the results would allow us to decide whether or not to eliminate the asymptomatic tree. When CLas infection is not validated throughout the process, further monitoring or a re-start of the survey would be followed if necessary.
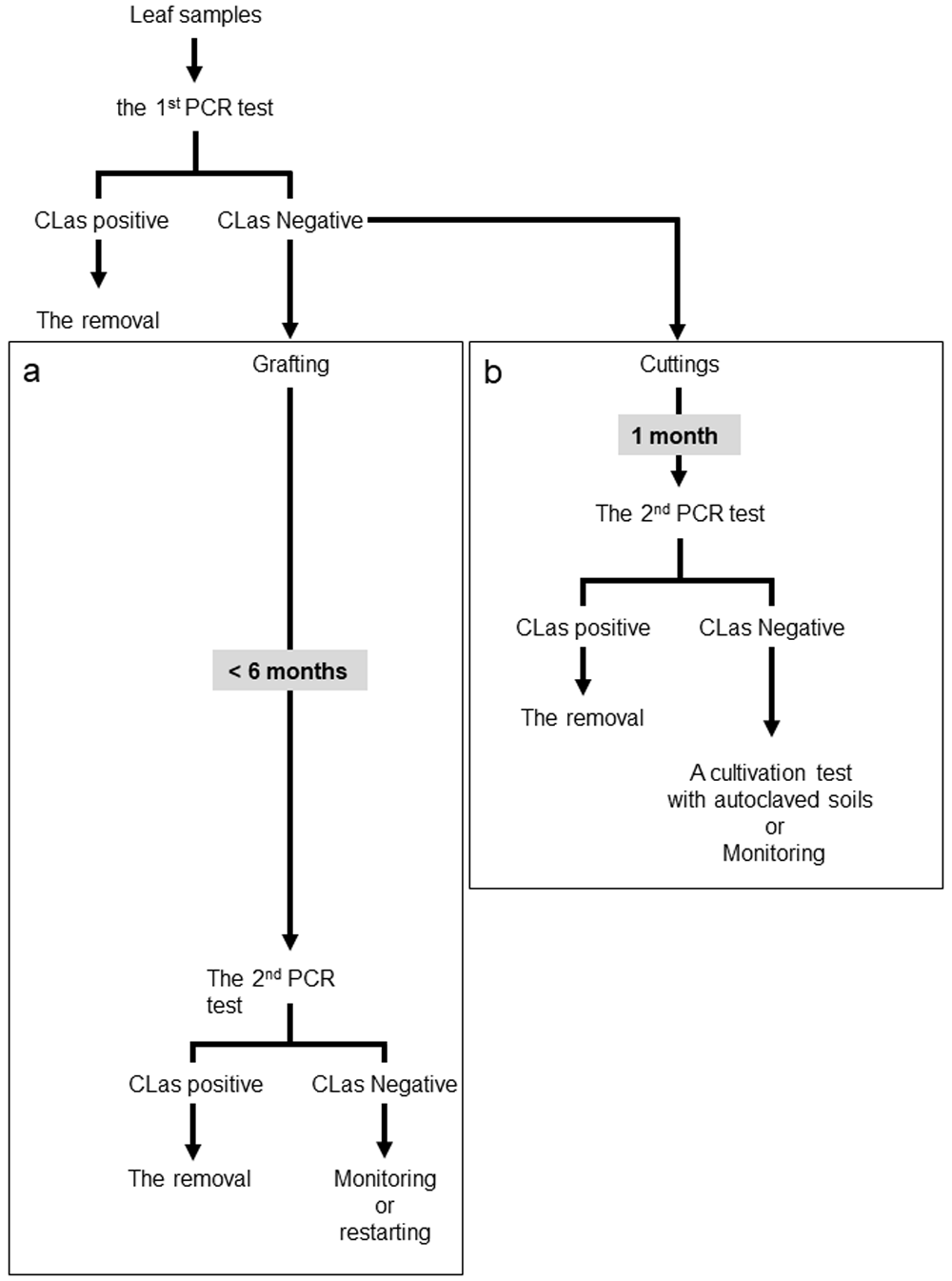
A new scheme for investigating latent infection of Candidatus Liberibacter asiaticus (CLas). Conventional (a) and new (b) approaches for validation of CLas infection in asymptomatic trees.
The techniques described in this paper are only applicable for citrus cultivars that root readily by cutting. These cultivars include lemon, trifoliate orange, murcot, Cleopatra mandarin, as well as the flat lemon that is common in the citrus greening-affected subtropical islands of Japan. For satsuma mandarin, oranges, and others that root poorly by cutting, propagation by air-layering would be needed. Exploration on media considering plant hormone formula and temperature conditions for promoting rooting from these cultivars is in progress in our laboratory.
The latent infection of CLas is detectable with the new detection method using cuttings. Our study revealed that the autoclaved nursery soils allowed for early detection of CLas. The new method using cuttings is time efficient in CLas surveying, in contrast to the conventional method which uses grafting. It is expected that the new method may provide opportunities for identification and validation of other ‘Candidatus Liberibacter’ spp. under quarantine conditions, as well as providing opportunities in monitoring surveys of citrus greening disease.
Figshare: Raw data for “Detection of CLas from roots recovered from cuttings” shown in Table 1. https://doi.org/10.6084/m9.figshare.13985021.v1 (Fujiwara, 2021a).
This project contains the following underlying data:
Figshare: Raw data for “A subsequent cultivation in different soil types using CLas-negative cuttings” shown in Supplementary Figures 2–6. https://doi.org/10.6084/m9.figshare.13985198.v1 (Fujiwara, 2021b).
This project contains the following underlying data:
Figshare: Supplementary Figures 1–7. https://doi.org/10.6084/m9.figshare.13466676.v1 (Fujiwara, 2020).
This project contains the following extended data:
- Figure 1. Schematic representation of the experimental design.
- Figure 2. A subsequent cultivation test for CLas detection in “Kenbyo” nursery soil using seedlings with no CLas detection in a primary PCR test.
- Figure 3. A subsequent cultivation test for CLas detection in “Sanyo composted bark” soil using seedlings with no CLas detection in a primary PCR test.
- Figure 4. A subsequent cultivation test for CLas detection in Acrisols using seedlings with no CLas detection in a primary PCR test.
- Figure 5. A subsequent cultivation test for CLas detection in Andosols using seedlings with no CLas detection in a primary PCR test.
- Figure 6. A subsequent cultivation test for CLas detection in “Kunigami Maaji” soil using seedlings with no CLas detection in a primary PCR test.
- Figure 7. A new approach for validation of CLas infection in asymptomatic trees.
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
No special permissions or quarantines were required for this study.
We thank Yuko Shimizu from Okinawa Prefectural Plant Protection Center. We also thank Katsuhiko Miyaji, Yoshihiro Ogawa, and Tomohiro Fukumoto from Kagoshima Prefectural Institute for Agricultural Development for supporting the soil collection. We are also grateful to Mariko Taguchi, Akane Sasaki, Hiroe Hatomi, and Yuko Fujii from NARO for their support throughout the experiments.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
No
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Kunta M, Viloria Z, del Rio H, Louzada E: Diverse DNA extraction methods and PCR primers for detection of Huanglongbing-associated bacteria from roots of ‘Valencia’ sweet orange on sour orange rootstock. Scientia Horticulturae. 2014; 178: 23-30 Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Citrus Pathology, Citrus Pathogen Detection, Horticulture
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Plant Pathology, some on the similar area studying of huanglongbing
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 29 Mar 21 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)