Keywords
image processing, matlab, multivariate image analysis, segmentation, interactive segmentation
This article is included in the NEUBIAS - the Bioimage Analysts Network gateway.
This article is included in the Software and Hardware Engineering gateway.
image processing, matlab, multivariate image analysis, segmentation, interactive segmentation
Modern imaging devices provide a wealth of data often organized as images with many dimensions, for instance 2D/3D, time and channel. This leads to new challenges in the exploration, visualization and processing of multi-dimensional data. In particular, multi-spectral images require the application of methods at the intersection of spectroscopy and image processing.
Matlab (The Mathworks, Natick, MA), is an efficient software solution for image and signal processing that provides native support for multi-dimensional arrays, a large number of image processing methods, and a great facility for adding custom developments. However, it lacks many features for facilitating the interactive interpretation of image data, such as a user-friendly visualization of multidimensional images, or the management of image meta-data (e.g. spatial calibration), thus limiting its application to bio-image analysis.
The ImageJ/Fiji software (Schneider et al., 2012) is an alternative that provides an intuitive graphical user interface for image exploration, and many image processing functions that take into account spatial or channel calibration. The basic functionalities can by extended by a large collection of plugins. The integration of custom developments is possible via macros or plugins, but this often requires advanced programming skills.
The ImageM software aims at providing a user-friendly interface for the interactive exploration, processing and analysis of multi-dimensional imaging data within the Matlab environment, while taking into account meta-data and facilitating the integration with custom algorithm developments.
ImageM strongly relies on Matlab’s Object-Oriented Programming. The core feature is an “Image” class that handles arrays up to five dimensions, corresponding to three spatial dimensions, the channels, and the time. It also encapsulates image meta-data such as spatial calibration, image name, or channel names.
The user interface relies on various image viewers adapted to the type (grayscale, color, label, multivariate) and the dimensionality of the image (Figure 1a). Interactive tools allow for quick image exploration, for example histograms, line profiles and 3D visualization. The architecture was kept modular to facilitate the inclusion of new features.
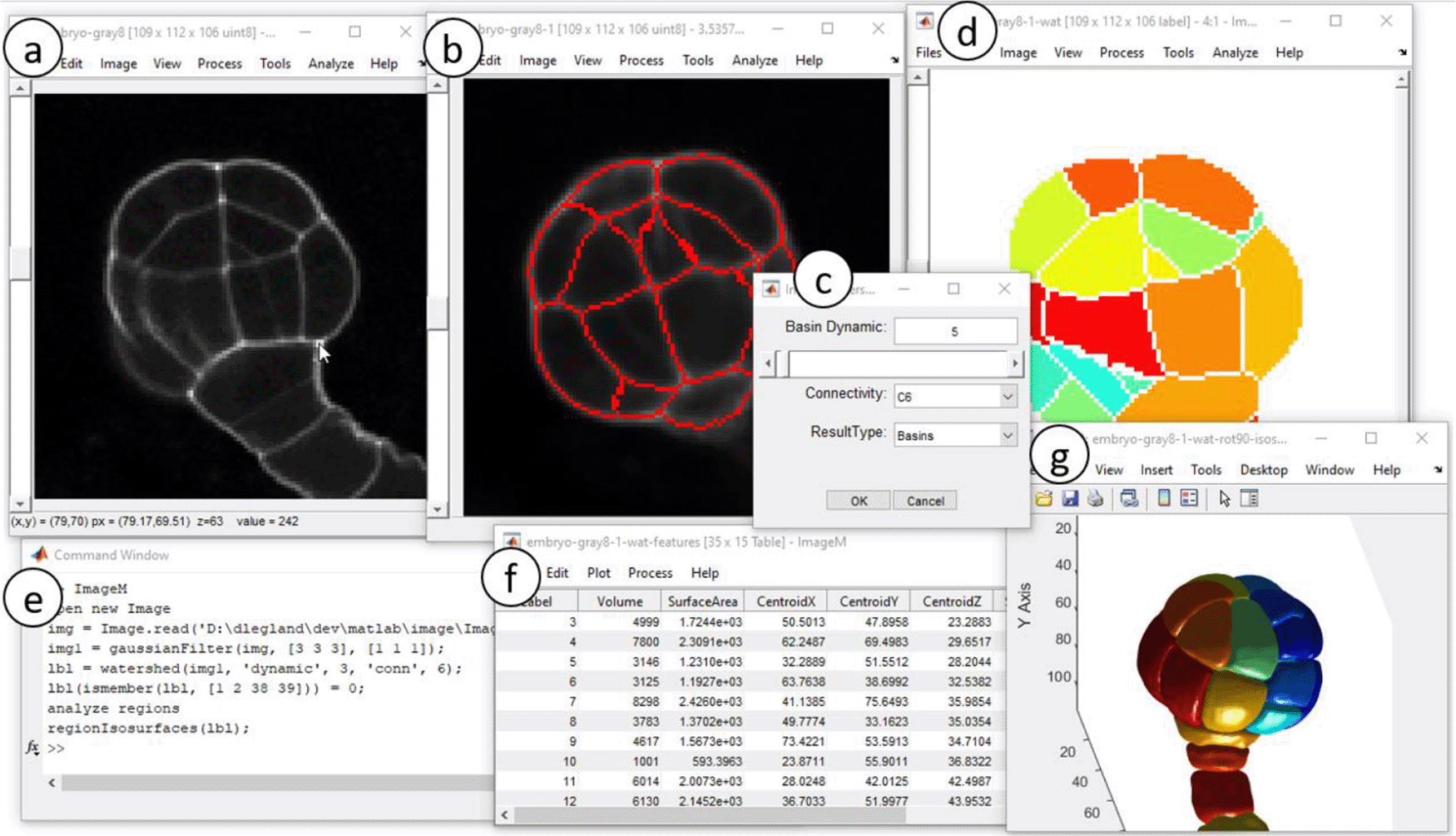
Original 3D image is visualized via adapted viewer (a). Image processing operators are applied (b), using contextual dialogs (c). Segmentation results are represented by label images (d,g). Analysis of the image results in table frame (f). Menu actions are transcripted into runnable Matlab commands (e). Original image courtesy of K. Belcram (Moukhtar et al., 2019).
To have the whole functionalities, Matlab (v2020a) is required, as well as the Image Processing and the Statistics toolboxes. The Gui Layout Toolbox (Sampson, 2021) is required to benefit from flexible image viewers. ImageM runs on a typical workstation. The application is launched by typing “ImageM” from the command line.
Menus provide quick access to common image processing operators, organized in a way to make them easily discoverable by a non-expert user. When necessary, intuitive dialogs allow for setting up parameters. Preview of the results facilitates the choice of relevant settings. Region of interest may be defined interactively to perform interactive measurements.
To facilitate usage during scripting and integration with custom developments, images can be easily imported from and exported to the workspace. Applying an operator results in a log entry that can be integrated into a script for quickly generating a workflow from the succession of operations.
ImageM integrates a large family of image processing operators (linear filtering, mathematical morphology, segmentation). Intuitive dialogs allow tuning of parameters and preview of the result, facilitating the choice of relevant settings (Figure 1c). The type of the result image is automatically inferred to provide the most adequate visualization (Figure 1d and e).
Segmentation algorithms result in binary or label images that can be used for quantitative analysis of regions. The spatial calibration of images is considered when available. Features can be visualized as geometric overlays on the image or exported to data tables (Figure 1f).
Multi-channel images or microscopy images contain pixels represented of several values. Multivariate image analysis provides a convenient formalism to analyze such images by taking account both the spatial and the spectral dimensions (Geladi & Grahn, 2006). ImageM can convert multi-variate images into data tables, allowing their exploration through multi-variate analysis tools such as Principal Component Analysis or k-means clustering (Figure 2). Resulting tables can be easily back-converted to images to facilitate the visual interpretation.
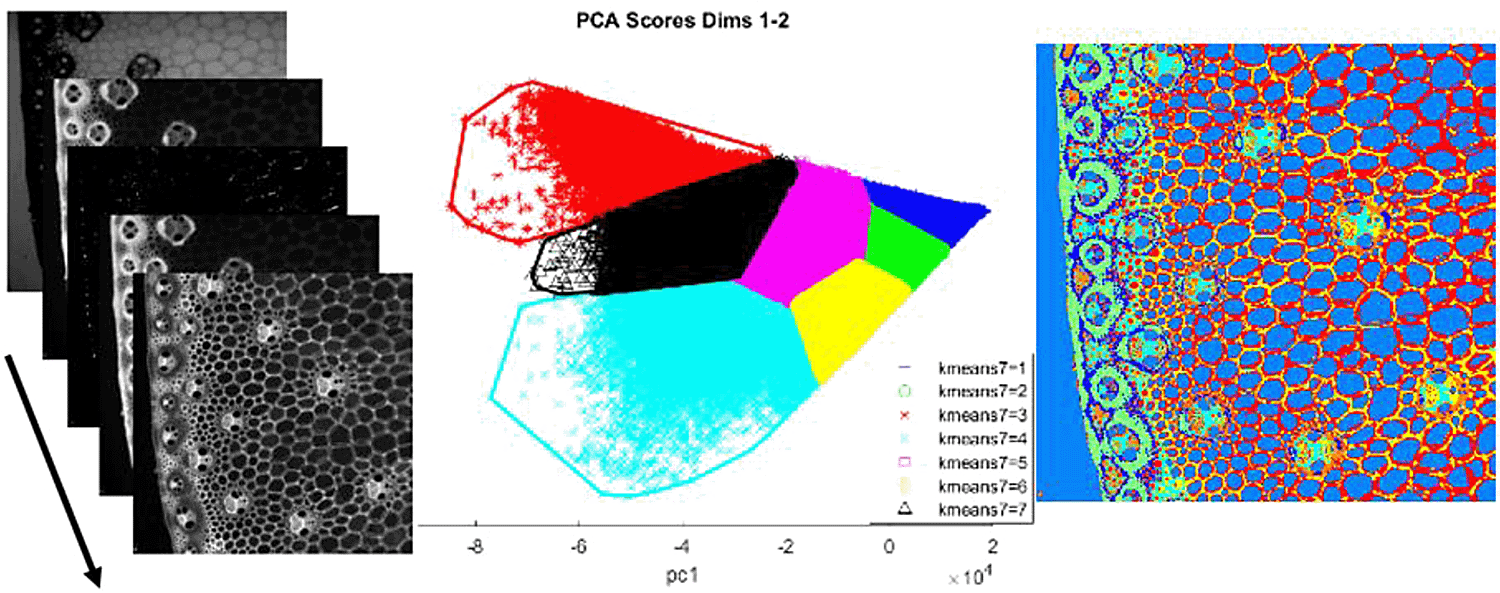
Left panel: original multivariate image (from Devaux, 2008). Middle panel: application of K-means clustering on principal components. Right panel: color representation of k-means classes.
ImageM can also run within the open-source platform, Octave, after some modification of the code (see Extended data (Legland & Devaux, 2021)).
We present here a use case describing image segmentation and region analysis of the 3D image presented in Figure 1. The Extended data (Legland & Devaux, 2021) contains the sample files, and more detailed use cases.
• First, run the application by typing “ImageM” from Matlab command line.
• In the menu, select “File->Open Image …” and choose the file “arabidopsis-embryo.tif”. This opens an image viewer for the 3D image.
• Reduce the noise in the image, by selecting “Process->Gaussian Filter …”. This opens a dialog.
• Choose value 1 for each of the “sigma” values, and click “OK”. The filtered image appears in a new viewer.
• Perform 3D segmentation of the cells. Select menu entry “Process->Extended Min Watershed …”. Choose a basin dynamic equal to 6, the “C6” connectivity, and result type as “basins”. This results in a new 3D label image in a new viewer, where each label is displayed with a different color.
• Some regions do not correspond to cells. They can be removed manually. Select the menu “Process->Replace Value(s) …” and fill in with “1, 2, 28, 29” (values can be identified by moving the mouse cursor and inspecting the status bar).
• Morphometric features can be computed from 3D label images, by selecting the menu “Analyze->Analyze Regions …”. After choosing the features and clicking “OK”, the selected features appear in a new data table frame that can be saved as a text file or exported to the workspace.
• The 3D representation of the Figure 1g can be obtained by selecting the menu entry “View -> Show 3D isosurface”, choosing a “smoothing radius” value equal to 1 and checking the options “Reverse Z-axis” and “Rotate Ox”.
The ImageM application proposes a convenient user interface for the visualization and exploration of multi-dimensional images within Matlab. It also provides basic support for regions of interests, the management of data tables. Its modular architecture should facilitate the future incorporation of new functionalities. The current work focuses on a better management of regions of interest, and on the inclusion of user plugins.
Packaged application available for direction download from: https://www.mathworks.com/matlabcentral/fileexchange/45847-imagem
Source code available from: https://github.com/mattools/ImageM
Archived source code as at time of publication: http://doi.org/10.5281/zenodo.4674326 (Legland, 2021).
License: BSD 2-Clause “Simplified” License
All data underlying the results are available as part of the article and no additional source data are required.
Zenodo: Extended data for the manuscript “ImageM: a user-friendly interface for the processing of multi-dimensional images with Matlab”, http://doi.org/10.5281/zenodo.4705240 (Legland & Devaux, 2021).
This project contains the following extended data:
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
This publication was supported by COST Action NEUBIAS (CA15124), funded by COST (European Cooperation in Science and Technology).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Partly
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Software engineering, software architecture, image visualization, image processing, image analysis.
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Computer Vision, Image Processing
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 30 Apr 21 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)