Keywords
Azoospermia, germ cell-specific protein, sperm production, sperm retrieval, TEX101
Azoospermia, germ cell-specific protein, sperm production, sperm retrieval, TEX101
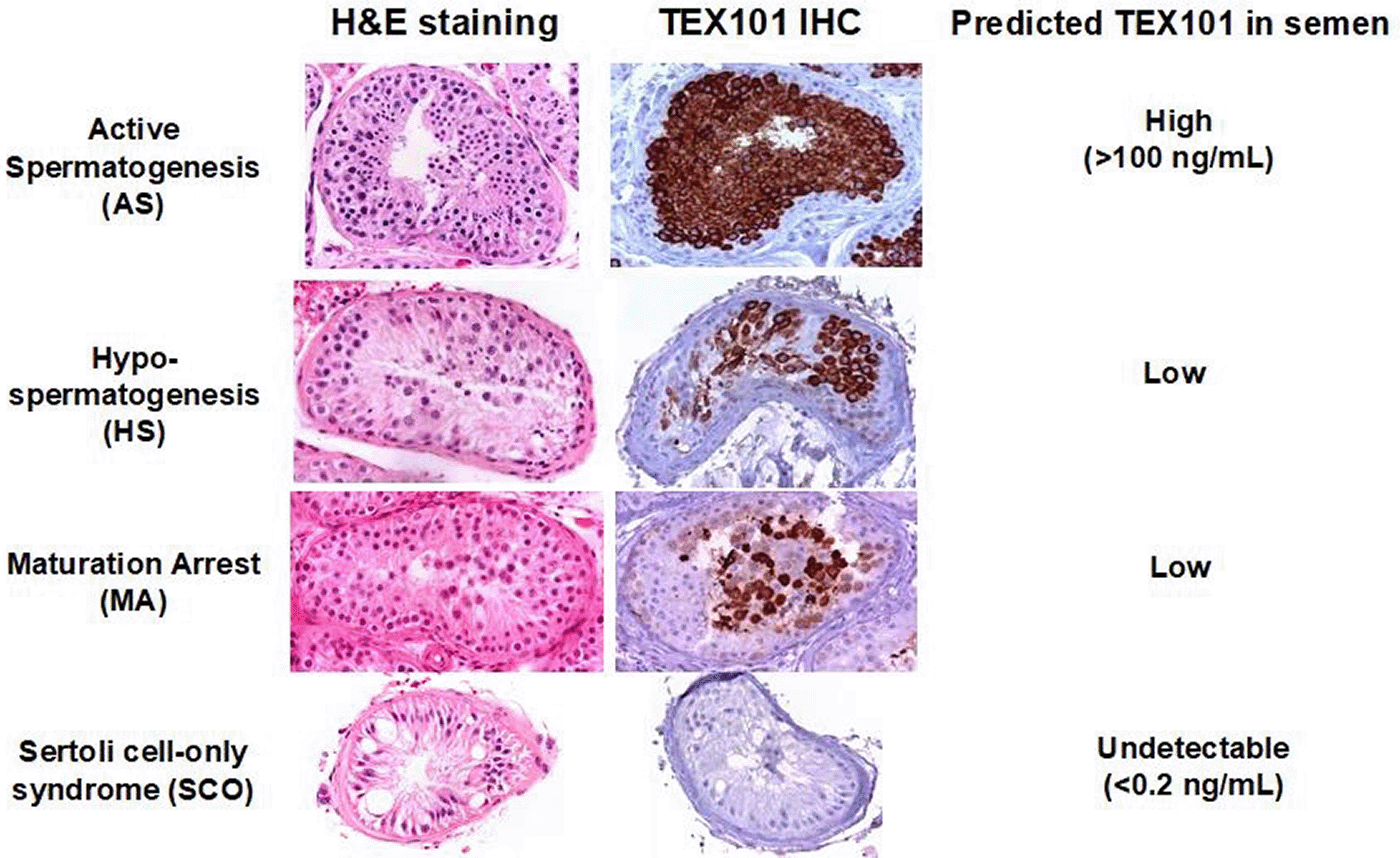
Of men with infertility, ~ 20% have azoospermia or no sperm in the ejaculate, with the majority (49–93%) of these men having spermatogenic failure (non-obstructive azoospermia (NOA)). While the term “failure” would seem to indicate complete absence of spermatogenesis, men with testicular failure have either reduced spermatogenesis (hypo-spermatogenesis), maturation arrest at either an early or late stage of spermatogenesis or a complete lack of spermatogenic cells noted with Sertoli-cell only syndrome.1–5
Until approximately 25 years ago, there were no available therapies that allowed men with NOA to have biologically related children. The advent of the era of intra-cytoplasmic sperm insertion (ICSI) with the capacity to inject a single sperm into an oocyte opened the door for successful therapies for men with very low sperm counts. This revolutionary therapy for couples with infertility was then extended to treat men with azoospermia, with sperm retrieved from the testis/epididymis/vas deferens being successfully used for ICSI.6–10 Testicular sperm extraction for men with NOA is now used world-wide with reported successful sperm retrieval rates of 30–70% and ICSI pregnancy rates between 19–50%.8,10–16
Non-invasive markers to predict the chances of a successful testicular sperm extraction technique are urgently needed. Several candidate clinical markers intended to predict the presence of sperm within the testis have been proposed including testis size, FSH, serum testosterone, inhibin B, genetic profiles (karyotype, Y-microdeletions) and blood flow patterns within the testis on Doppler studies.15–19 Unfortunately, none have demonstrated consistent clinical reliability compared with an in-depth testicular tissue analysis and sampling using the surgical microscope. Even the pathology on a previous diagnostic testicular biopsy (usually a randomly sampled tissue biopsy) does not definitively predict whether sperm are present or absent throughout the remaining of testis among men with non-obstructive azoospermia.7,8,11,13,14,18,20,21
In previous work from our laboratory, using liquid chromatography-tandem mass spectrometry (LC-MS/MS), we identified two seminal proteins, epididymis-specific protein ECM1 and germ cell-specific protein TEX101 that were able to differentiate obstructive versus non-obstructive azoospermia with 100% specificity and 81% sensitivity.22–27 Immunohistochemical staining of testicular tissue revealed that TEX101 is found in the primary spermatocytes and more mature sperm forms, but not in spermatogonia, Sertoli or Leydig cells.24,27 In addition, according to Human Protein Atlas, TEX101 protein is expressed in germ cells only, and not in any other of 75 tissues and cell types studied (http://www.proteinatlas.org/ENSG00000131126).
This background information suggests that TEX101 concentrations should be non-detectable in the semen of men with diffuse Sertoli-Cell-only syndrome and maturation arrest at the spermatogonial stage of development (early maturation arrest) (Figure 1). Non-detectable TEX101 could be useful to predict sperm retrieval failure.
In our previous work, we identified TEX101 as a potential marker for sperm production in men with NOA.23,24 This work used mass spectrometry (MS) to measure the abundance of TEX101 in the semen of men with NOA but was limited due to fairly high limit of detection (~5 ng/mL) and the cost of MS, making routine use for diagnostic testing impractical.
We have produced several monoclonal antibodies (mAbs) against human TEX101 and paired two mAbs to develop a highly sensitive, reproducible and low-cost ELISA assay with detection limit of < 0.2 ng/mL.27–29 With our recent development of a highly sensitive, reproducible and inexpensive ELISA assay for TEX101, we are now able to better study how accurately semen TEX101 levels predict sperm retrieval for men with NOA.
This study was approved by the research ethics boards of both the Sinai Health System and the Weill Cornell Medical Center and all participants provided written informed consent for the use and publication of the patient’s data.
Between Jan 2016 to Jan 2019, patients were recruited prospectively from the specialist male infertility clinics at the University of Toronto and Weill–Cornell University. Semen samples were collected prior to surgery with the last samples being collected in June 2019. Men who were eligible for the study (presumed NOA with a minimum of two semen assays confirming azoospermia/cryptozoospermia and clinical parameters compatible with NOA) who were planning on proceeding with sperm retrieval were approached to enter the study. Since no additional semen samples were required and the surgery offered to the men was not changed by participation in this study, non-participation in the study was uncommon. To avoid bias, all potential patients were recruited.
The semen specimens were collected by masturbation into a sterile collection cup either at home or at the clinic. Following liquefaction (~1 hour at room temperature) a semen analysis was performed, then the semen samples were immediately centrifuged at 4oC and the supernatant (seminal plasma, SP) was frozen at −80oC. Whenever possible, semen samples were provided monthly for up to 3 months.
The production and selection of mouse monoclonal anti-TEX101 antibodies has been previously described.29,30 For the immunoassay development, white 96-well ELISA plates were coated with 400 ng/well of mouse monoclonal anti-TEX101 antibody 34-ED-629.3 in 50 mM Tris buffer, pH 7.8. Plates were washed (2×) with 0.05% Tween 20 in 20 mM Tris, 150 mM NaCl, pH 7.4). TEX101 calibrators and diluted samples in assay buffer containing 60 g/L BSA, 20 mg/L Mouse IgG, 100 mg/L goat IgG,1 g/L Bovine IgG and 0.25 M KCl in 5 0mM Tris with 0.05% Tween 20, pH 7.8, were added into each well (100 μL/well) and incubated for 2 h with gentle shaking. Plates were then washed (3×) with the washing buffer. A biotinylated mouse monoclonal anti-TEX101 antibody 34-ED-470.3, diluted in a solution containing 60 g/L BSA, 20 mg/L mouse IgG, 100 mg/L goat IgG, and 1 g/L bovine IgG in 50 mM Tris with 0.05% Tween 20, pH 7.8, were added (25 ng of antibody per 100 μL of solution per well) and incubated for 1 h. Plates were washed (6×) and alkaline phosphatase-conjugated streptavidin was added in the wells (100 μL per well). Incubation was for 20 min at RT with gentle shaking, followed by a final wash (6×). Diflunisal phosphate (DFP) solution was prepared in substrate buffer (0.15 M NaCl, 1 mM MgCl2 in 0.1 M Tris, pH 9.1), added on the plate (100 μL per well) and incubated for 10 min at RT with gentle shaking. Subsequently, the developing solution (1 M Tris, 0.15 M NaOH, 2 mM TbCl3, and 3 mM EDTA) was added on top and mixed for 1 min. Time-resolved fluorescence was measured with the Wallac EnVision 2103 Multilabel Reader (Perkin Elmer).
The calibrators were prepared by pooling several seminal plasma samples. TEX101 concentration in the pool was calculated using a quantitative SRM method, described elsewhere.30 Aliquots (volume of 20 μL each) of the pool were prepared and stored at −20 °C.
To assess the assay’s linearity, serial dilutions of the calibrator sample were prepared in a two-fold dilution step (ranging from ~7 ng/mL to ~0.05 ng/mL) and added on ELISA plate. Each dilution was analyzed in four replicates. For within-run and total precision, three levels of TEX101 concentration in the calibrator sample were used (mean 0.0770 ng/mL, 0.322 ng/mL, and 3.70 ng/mL). Each concentration was analyzed in eight replicates per run (plate), in two runs per day, over three days.
To assess assay’s linearity and within-run and total precision, DataInnovations software was used. The open-source software HYPERLINK “http://www.gnumeric.org/” HYPERLINK “http://www.gnumeric.org/” Gnumeric (Gnumeric, RRID:SCR_018462) may also be used in place of EP Evaluator to preform similar analyses.
TEX101 ELISA assay limit of detection (LOD) is 0.015 ng/mL. Since seminal plasma samples are diluted 10-fold before analysis, the LOD for patient samples is 0.15 ng/mL; rounded to 0.2 ng/mL.
TEX101 concentration is stable in SP samples stored at 4°C and 22°C over a period of at least 7 days. Samples were run in triplicate.
Sperm retrieval was performed using a standard technique as described by Schlegel in 1999.8 If any sperm was identified during or after the procedure, this was considered positive sperm retrieval.
From previous studies on semen TEX101, we had shown that men having a Sertoli Cell Only pattern (lacking germ cells) would have undetectable TEX101 in the semen, while those with complete spermatogenesis would have detectable TEX101 levels.23,24 Since successful sperm retrieval rates for men with NOA are reported to be between 30-70%, we calculated the sample size required of 54 based on an estimated successful sperm retrieval rate of 50% in those with detectable TEX101 in the semen vs 15% for those with undetectable TEX101 in the semen (power of 0.8 and alpha of 0.05).8,10–16 We aimed to recruit an additional 25% to account for drop-outs and incomplete information.
Comparison of binomial results was performed using Fisher’s exact testing using Microsoft Excel (RRID:SCR_016137; An open access alternative is Google Sheets (RRID:SCR_017679)), and 95% CI were estimated using an online calculator.
Two mouse monoclonal antibodies (34-ED629.3 and 34-ED-470.3) targeting different epitopes of native TEX101 were used to develop a sensitive immunofluorometric assay. The new assay allowed a significant increase of ELISA signal with no sample pre-treatment required.30
The limit of quantification (LOQ) of the immunoassay was set at the TEX101 concentration showing CV≤20%, which was 0.015 ng/mL. The limit of detection (LOD) of the assay was defined as LOD=LOQ/3, which was 0.05 ng/mL. The linear range of the assay spanned from 0.015 ng/mL to 7.2 ng/mL (slope 0.966; intercept −0.0010) (Figure 3). Within-run (n=3 different levels, each in eight replicates) and between-run (n=3 different levels, each in eight replicates per run) precision assessed over 3 days were <6 and 9%, respectively (Table 2).
Recovery was assessed by spiking a seminal plasma pool (of TEX101 concentration: 0.27 ng/mL and 1.1 ng/mL) into four seminal plasma samples with known TEX101 concentration (measured by a quantitative SRM method). Recovery of TEX101by this assay ranged from 93 to 104%.
A total of 69 men (mean age 35±4.6 years) with NOA, considering undergoing surgical sperm retrieval, were enrolled in this study. Karyotype analysis revealed non-mosaic Klinefelter’s syndrome in four out of 69, a deletion of Y(q11.22) in one out of 69 and 64 out of 69 with normal karyotypes.
All of the 69 men enrolled in the study provided at least one semen sample for the TEX101 assay, with 37 providing more than one sample for analysis at least one month following the initial testing. Surgical sperm retrieval was performed in 65 out of 69 men, of whom 60 had normal karyotypes.
Semen TEX101 concentration was less than the limit of detection (LOD <0.2 ng/mL) for 25 out of 69 initial semen specimens tested, with the median concentration of the remaining 44 semen samples being 0.98 ng/mL (IQR 0.35–126 ng/mL: range 0.2–23,102 ng/mL).
A total of 37 out of 69 men had more than one semen sample tested. Of these 37 men, eight had consistent TEX101<LOD, 20 had consistent TEX101>LOD and nine had variable TEX101 (above and below LOD).
The results of the surgical sperm retrieval procedures and the semen TEX101 levels were available for a total of 65 men. Overall, sperm was found in 23/65 sperm retrieval procedures.
While there were only four men with Klinefelter’s syndrome, TEX101 semen levels were low in this group with 3/4 having semen TEX101 <0.2ng/mL but sperm retrieval rates were high (75%).
Out of the 65 men above, a total of 60 men had a normal karyotype. Of these 60 men, zero out of 20 (0%) of those with semen TEX101 <0.2 ng/mL (TEX101 measure on the semen sample provided closest to the date of the surgical sperm retrieval if more than one sample provided) had sperm identified during the surgical sperm retrieval procedure, while 20 out of 40 (50%) of those with semen TEX101 >0.2 ng/mL had sperm retrieved with surgery (significantly different, Fisher’s exact test, p < 0.05) (Table 1).
| TEX101 ng/mL | Sperm identified (%) | 95% CI |
|---|---|---|
| <0.2 ng/mL | 0/20 (0%) | 0–17% |
| >0.2 ng/mL | 20/40 (50%)* | 34–66% |
| Any concentration | 20/60 (33%) | 22–47% |
| Within-run | Between run (3 Days) | |||
|---|---|---|---|---|
| TEX101 levels (ng/mL) | SD1 | CV (%) | SD | CV (%) |
| 0.0770 | 0.003 | 4.2 | 0.006 | 8.5 |
| 0.322 | 0.011 | 3.5 | 0.015 | 4.7 |
| 3.70 | 0.20 | 5.4 | 0.28 | 7.5 |
This new study documents that TEX101 semen concentrations predict sperm retrieval outcomes for men with NOA and normal karyotypes. None of the men with TEX101 semen concentrations below the assay’s LOD had sperm found, while sperm was found in 50% of those with semen TEX101 levels above the assay’s LOD.
We have previously shown that the concentration of two semen protein markers, TEX101 and ECM1 differentiate with high specificity and sensitivity NOA from OA.24 ECM1 is an epididymal mono-specific marker which was below detection limits for men with OA, but present in the semen in detectable amounts in men with NOA. TEX101 is a germ cell mono-specific marker found on sperm, round spermatids, and spermatocytes, but not in spermatogonia, Sertoli cells or Leydig cells.24,27–29 Since TEX101 is germ cell mono-specific, it follows that semen TEX101 levels will be non- detectable in men with obstructive azoospermia and should also be non-detectable in men with Sertoli cell-Only syndrome and those with maturation arrest at the spermatogonia stage.24
It is important to note that the majority of the small number of studied men with Klinefelter’s syndrome had undetectable TEX101 semen levels but sperm retrieval rates remained high. The lack of detection of TEX101 of men with Klinefelter syndrome may reflect the focal nature of spermatogenesis that is commonly present in these men and/or sclerosis of the tubular lumen.
These results should have important implications in how we test and manage men with azoospermia. The information from the semen concentrations of ECM1 and TEX101 will differentiate OA from NOA with high specificity and sensitivity (Figure 1).27 For those with NOA, normal karyotype and undetectable semen TEX101, the test predicts that these men are very likely to have uniform Sertoli cell-only Syndrome or early maturation arrest with low/zero success rates with sperm retrieval (0%: 95% CI 0–17%). We have proposed an algorithm to diagnose the category of azoospermia and the expected outcomes of the sperm retrieval procedure based upon this study and our previously published work (Figure 2). This information should help clinicians counsel couples about their expected success rates as they manage azoospermia.
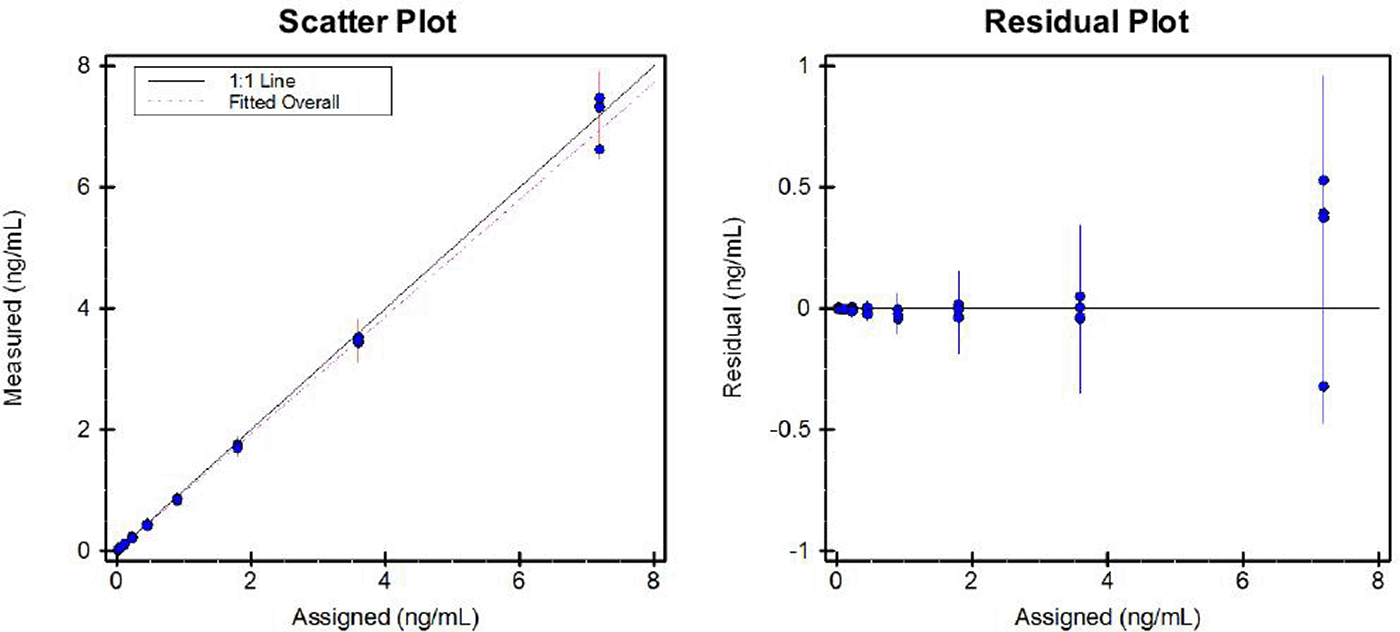
Scatter plot illustrating the assigned (theoretical) and measured TEX101 concentration in serial dilutions of seminal plasma calibrator sample analyzed in four replicates (left); Residual plot showing the assigned (theoretical) TEX101 concentration and the residual concentration (the difference between the measured and the assigned TEX101 concentration) (right).
Interestingly, we did notice variability of semen TEX101 levels, with some men varying between detectable and undetectable TEX101 levels. It is possible that this variability indicates some underlying changes in spermatogenesis (similar in concept to the known variability in sperm counts in men with sperm in the ejaculate), so potentially TEX101 levels may be used in the future to time surgical sperm retrievals for men with NOA to optimize success rates.
There are two important caveats to consider with the results of this study. The observed sperm retrieval failure rates for those with semen TEX101 concentration <LOD was 100%, with a 95% CI of 83–100%. This study should not be used to conclude that men with TEX101 < 0.2 ng/mL universally have no sperm production, but should suggest that the prognosis for sperm retrieval is very poor for this group of men. Further studies on more men from different centers are required to provide a more accurate estimate of the negative predictive value of this test, as techniques for, and success of, sperm retrieval may vary at different centers.
Secondly, the men seen and the type of surgery performed at the Toronto and Weill–Cornell centers may not be representative of the general male infertility practice. A different population of men with NOA will change the mean TEX101 concentration and fraction with TEX101 <LOD, while the type of surgery alters the success rates of the sperm retrieval. Further studies on men with NOA from different male infertility clinics are needed.
Keith A. Jarvi had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
University of Toronto Dataverse: Underlying data for ‘Semen biomarker TEX101 predicts sperm retrieval success for men with testicular failure’. https://doi.org/10.5683/SP2/DQYDQW.31
The project contains the following underlying data:
• TEX 101_Cornell-Jun 1 2021.tab (Cornell sample with their TEX101 concentration and if sperm was identified during mTESE)
• TEX101_Sinai-Mar 11, 2019.xlsx (Sinai sample with their karyotype, TEX101 concentration and if sperm was identified during mTESE)
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: More clinical data and correlations are needed as discussed before
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Drabovich AP, Dimitromanolakis A, Saraon P, Soosaipillai A, et al.: Differential diagnosis of azoospermia with proteomic biomarkers ECM1 and TEX101 quantified in seminal plasma.Sci Transl Med. 2013; 5 (212): 212ra160 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Male reproduction
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 14 Jul 21 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)