Keywords
Casein, Enzyme activity, Leaf extract, Moringa Oleifera, Plant-derived proteases, Protein purification.
Casein, Enzyme activity, Leaf extract, Moringa Oleifera, Plant-derived proteases, Protein purification.
All organisms contain proteases that hydrolyze peptide bonds in order to maintain systemic homeostasis and for its normal growth and development1,2. Proteases derived from plants, animals and microbes find wide industrial applications including in the leather, food, brewery and pharmaceutical industry2–4 corresponding to approximately 60% of the total worldwide enzyme sales5.
Moringa oleifera is one of the best known medicinal plants widely distributed in the tropical regions6. It contains a mixture of several hydrolytic enzymes, in which proteases are the key enzymes reported to show pharmacological activity7. We attempted to investigate the protease activity of aqueous extracts of Moringa oleifera leaf. Here, we have isolated and purified the protease from Moringa leaves and carried out enzyme kinetics study and find that the protease exhibited optimal caseinolytic activity in alkaline pH.
Mature Moringa Oleifera leaves were collected from a plant located near TIU campus, Salt Lake Kolkata and crushed along with 20mM phosphate buffer (pH 7.5) and 0.1% tween 20 detergent and protease cocktail inhibitor followed by centrifugation with plastocraft table top refrigerated centrifuge machine (Rota 4RV/FM) at 10000 rpm for 10 mins at 4°C. The crude soup was mixed with 40% ammonium sulphate to obtain the protein precipitate, which was then dissolved in 20 mM tris buffer for further evaluation.
The total protein content of the solutions at different stages of protein purification was determined by Bradford methods8 using Sigma’s Bradford reagent (B6916). In this assay, a series of BSA standard solutions (0.1 – 1.2mg/ml) were used to prepare the standard curve. Bradford assay was performed by adding 1 mL of Bradford reagent to 20 μl of each standard solutions or unknown solution, and homogenized by using vortex mixer. The samples were incubated in dark conditions for 10 minutes and the absorbance was read at 595 nm.
We performed sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) using 12% resolving and 5% stacking gels for separating proteins. We followed the Laemmli’s method9 for gel electrophoresis. The samples were mixed with equal volume of gel loading buffer and heated at 95°C in dry heating bath for 2 mins. The electrophoresis process was run with 90 V for first 10 mins and then run at 150 V with Biorad mini protean gel electrophoresis system. After complete run the gel was stained with Coomassie Brilliant Blue. We have used protein marker (10kD to 250 kD) from GCC biotech (Pre-stained protein marker GCR-P4B) for determination of molecular weight. We imaged the gels in Biorad gel documentation system. Acrylamide, bis acrylamide, Tris and TEMED (T9281) are from Sigma Aldrich. Coomassie Brilliant Blue R250 (93473) and Ammonium per sulphate (28575) was from SRL (Sisco Research Laboratories).
Dialysis: The pellet dissolved in Tris buffer as obtained above was then dialyzed in 3.5cm/ml dialysis tubing (SIGMA Aldrich D6066 overnight in a magnetic stirrer by immersing the tubing in a buffer containing Tris (pH 8) and phenylmethylsulfonyl fluoride (PMSF) SRL, which was repeated thrice for complete exchange of buffer.
Diethylaminoethyl (DEAE) cellulose ion exchange chromatography: The protein sample was loaded in the DEAE cellulose (SIGMA Aldrich 30477) column. Ion exchange column chromatography was carried out by using an assembly of Biorad’s Econo pump model EP-1, UV monitor and chart recorder from Atto, Japan and Biorad’s fraction collector model 2110. A gradient of 0.05 M to 0.5 M NaCl was used to elute the protein from the column. The gradient was run for 150 min with a flow rate of 1ml/min. Optical density (OD) of all the fractions were taken at 280 nm with Schimadzu 2401 UV Vis Spectrophotometer.
Samples at different stages of purification were tested for albuminolytic property of protease by using BSA SIGMA as substrate. BSA digestion was performed at 37°C and pH 7.5 for 1 hour. Further, each of the samples were mixed with protein gel loading dye in 1:1 ratio and loaded in SDS PAGE and the gel was imaged with Biorad gel documentation system.
In this assay, β-casein was used as substrate. If protease digests casein, the amino acid tyrosine is liberated along with other peptide fragments. Folin’s reagent reacts with free tyrosine to generate a blue colored product, which is quantifiable and measured as an absorbance value on the Schimadzu UV 2401 spectrophotometer at 660 nm. A tyrosine standard calibration curve is constructed to determine the amount of tyrosine released after the proteolytic activity. A series of tyrosine standard solutions at different concentrations (5 – 50 μg/mL) were prepared from the 0.18mg/mL L-tyrosine stock solution with deionized water. L-tyrosine was purchased from Himedia, Fohlin’s reagent was obtained from SRL and β-casein from SIGMA.
We have assayed the protease activity in terms of caseinolytic activity with plant leaf extracts at different stages of purification (crude soup is the initial supernatant after homogenization and centrifugation, 40% ammonium soup is the phosphate dissolved pellet after 40% ammonium sulphate fractionation and pooled soup is the final collection of pure fractions came from DEAE cellulose column). All the three samples were dialysed to remove protease inhibitor and EDTA before the protease assay. The protease activity of pure protein was examined at different pH range 4–9 and temperature range 4–70°C.
The enzyme activity assay for protease was conducted with different concentrations of β-casein as substrate, at pH-8 in 37°C respective optimum conditions as determined with the previous experiments described above (optimum temperature and pH conditions). Here the substrate concentration (β-casein) varied in the range (0.81, 1.6, 2.4, 4.03, 5.2) mg/ml keeping the enzyme concentration fixed.
Moringa oleifera leaves are reported to contain protease but there are no detailed studies on the purification and kinetic parameters of the enzyme. Here, we obtain partially purified protease from the aqueous extract of the leaves by ion exchange chromatography such that in anion exchange the proteins show a peak at 280 nm implying a positively charged protein.
The protein concentration from mature Moringa oleifera leaves at various stages of purification is shown in Table 1, which was purified by DEAE cellulose ion exchange column chromatography. The chromatogram for purification is shown in Figure 1A. The purified fractions were observed in 12% SDS PAGE (Figure 1B). The protein was of 51 kDa according to molecular weight markers.
| Samples | Protein concentration(mg/ml) |
|---|---|
| Crude | 0.56 |
| 40% | 0.55 |
| Pooled | 0.22 |
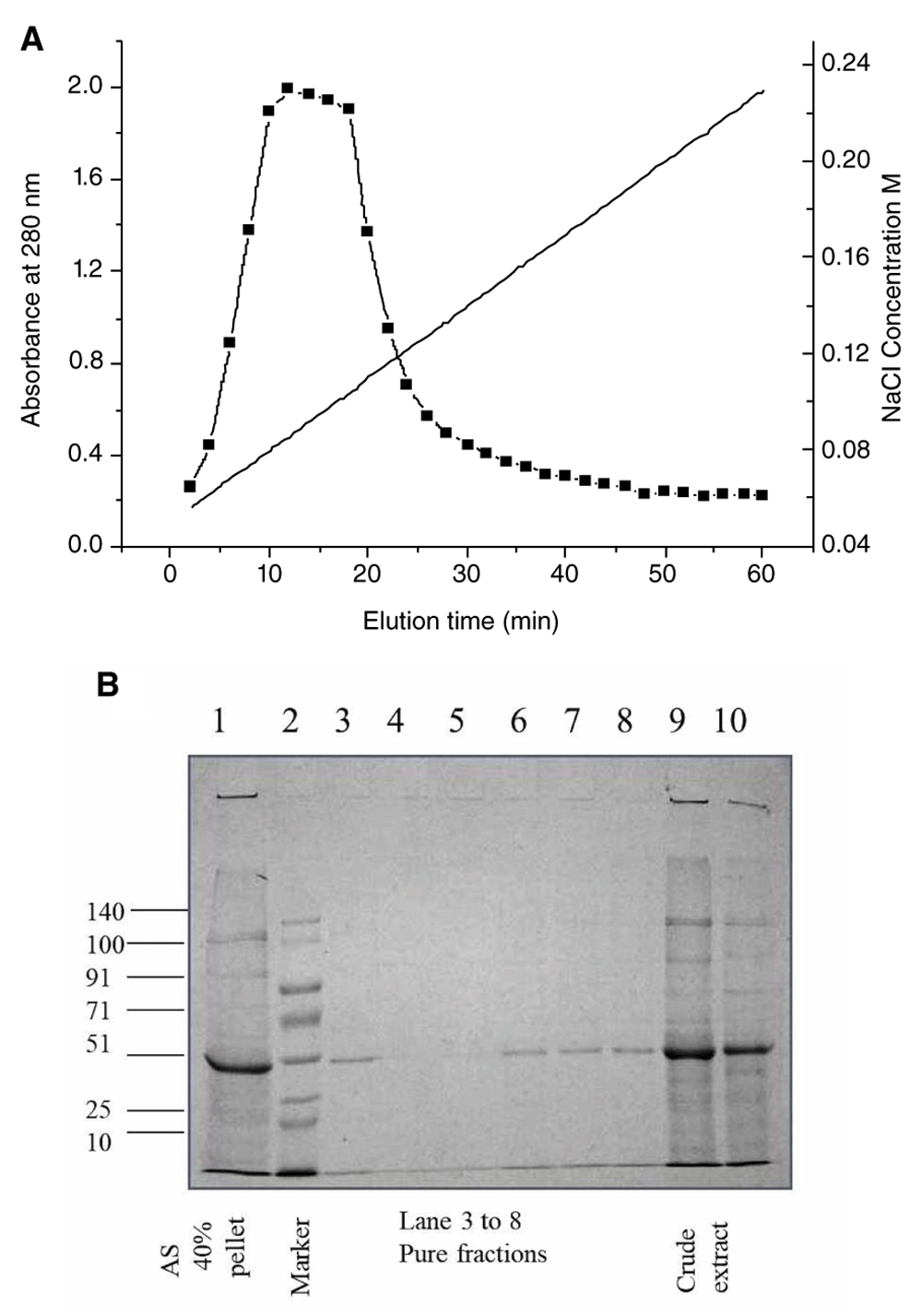
A. Chromatogram for the purification of protein from Moringa oleifera shows the elution time versus absorbance at 280 nm and the corresponding NaCl gradient profile (ranging from 0.04M to 0.25M) for maximal elution B. SDS PAGE of the crude extract and fractions after purification by DEAE cellulose ion exchange chromatography. Lane 1 shows extract after 40% ammonium sulphate precipitation, lane 2 shows the prestained molecular weight marker from GCC biotech marking 140, 100, 91, 71, 51, 25 and 10 kDa bands, lanes 3 to 8 represent fractions after column purification, lanes 9 and10 show the bands from crude leaf extract.
Results from Figure 2 shows that both crude extract and 40% ammonium sulfate fractionated sample possesses protease activity and is able to produce fragments of BSA (lane 5, 6 and 9).
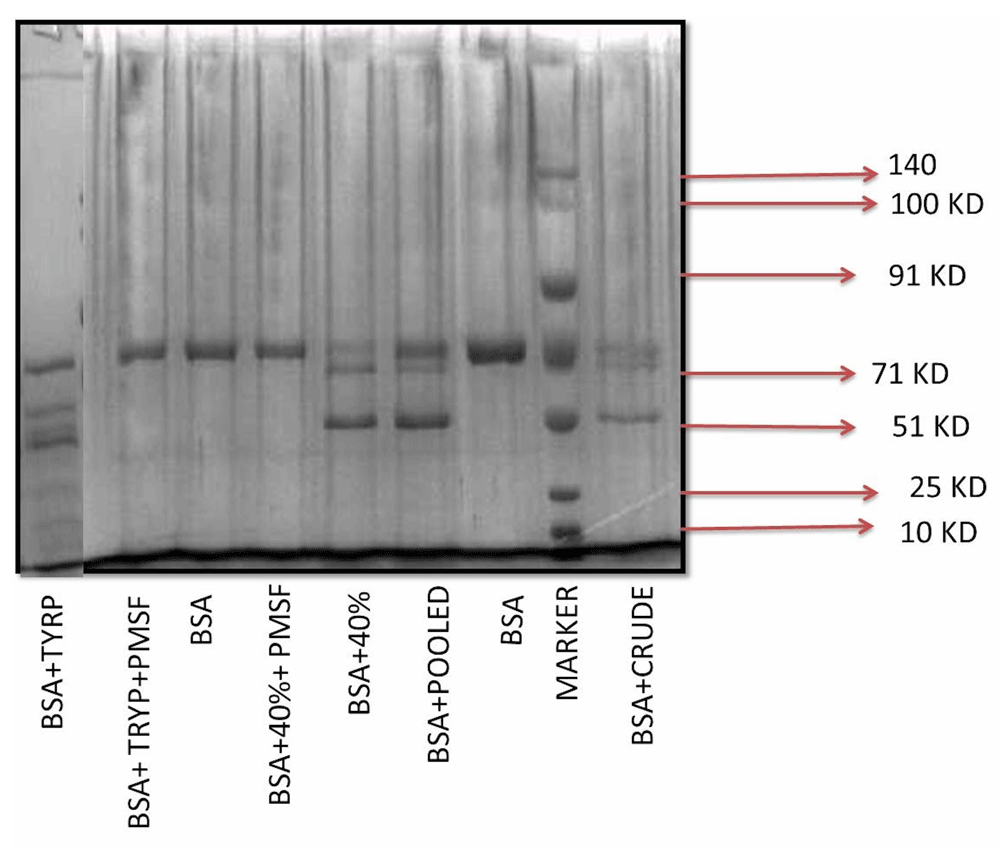
Lane 1 is BSA and trypsin, lane 2 is BSA + trypsin + PMSF, lane 3 is BSA, lane 4 is BSA +40% Ammonium Sulphate cut + PMSF, lane 5 is BSA + 40% Ammonium Sulphate cut, lane 6 is BSA + Pooled pure protein, lane 7 is BSA, lane 8 is prestained molecular weight marker from GCC biotech showing 140, 100, 91, 71, 51, 25 and 10 kDa bands and lane 9 is BSA + crude leaf extract.
UV-vis absorption spectra of the pure protease were shown in Figure 3. A single peak at 280 nm can be observed for the pure protein.
In both crude extract and purified protein, protease activity was measured as described in methods. Reactions in different pH 4, 5, 6, 7, 8 and 9 were done (Figure 4). The results showed maximum activity at the pH 8.0. Therefore, the enzyme is an alkaline protease.
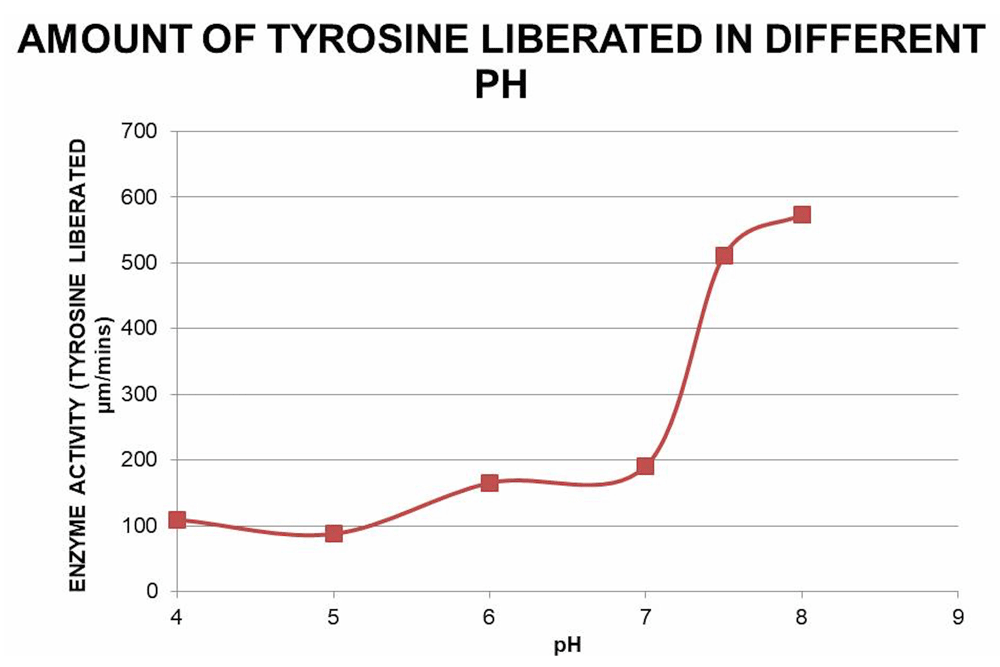
Protease activity of the pooled pure fractions on β-casein degradation is plotted against different pH (4, 5, 6, 7 and 8) at 37ºC. Free tyrosine liberated due to β-casein degradation was monitored with Folin-Ciocalteau reagent at 660 nm and the corresponding amount was measured from the tyrosine standard curve.
The protease assay with β-casein as substrate was performed at a range of temperatures; 4°C, 25°C 37°C, 55°C and 70°C (Figure 5) according to the methods described above. The enzyme activity was found to be maximum at 37°C.
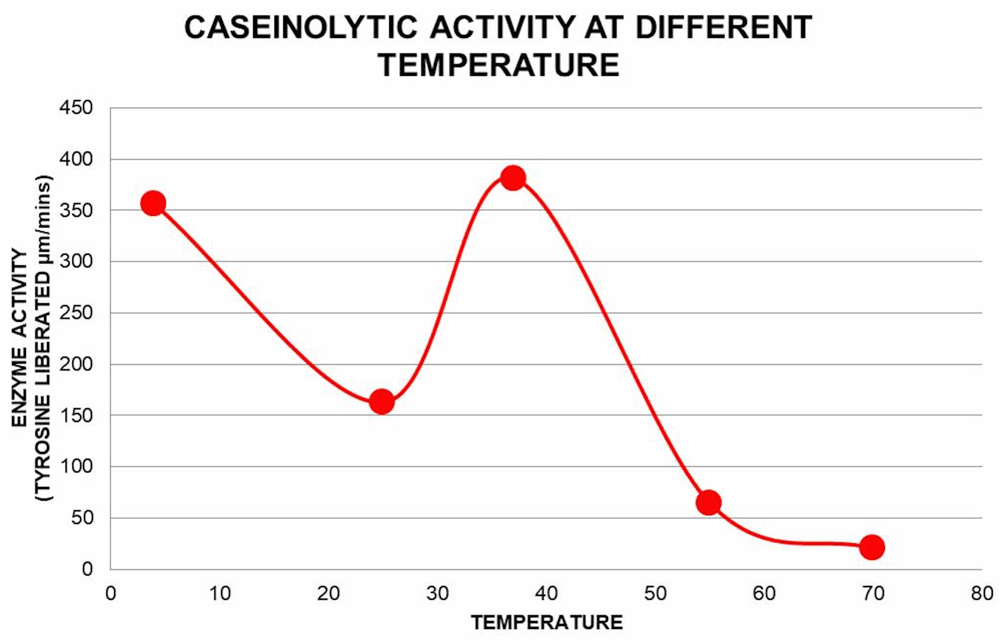
Protease activity of the pooled pure fractions on β-casein degradation is plotted against different temperature (4, 25, 37, 55 and 70°C) at pH 8. Free tyrosine liberated due to β-casein degradation was measured as described earlier.
Specific activity of the protease was calculated by enzyme activity from the protease assay using β-casein as substrate and the total protein content of the protease solution. We can see a large increase in specific activity after the final purification (Figure 6).
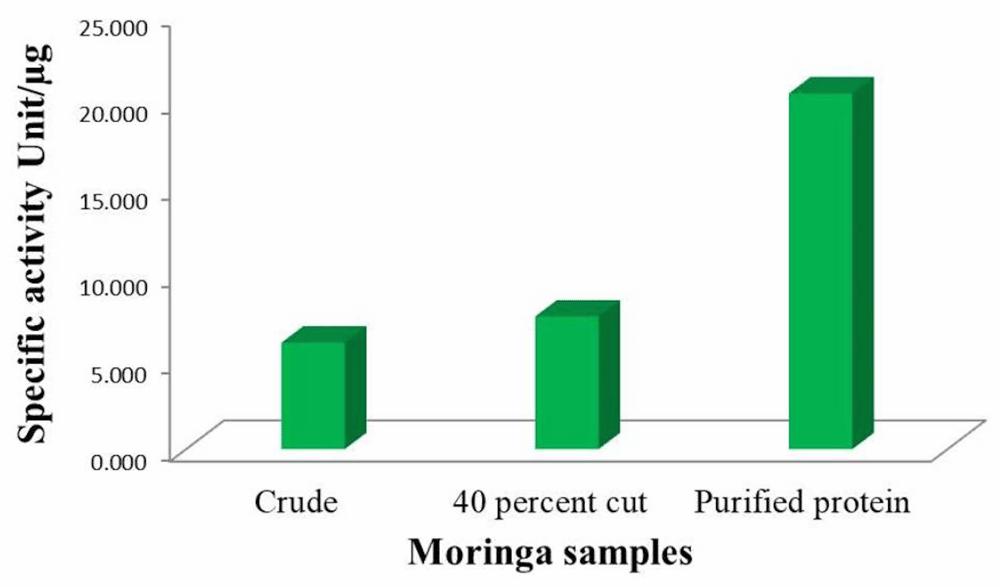
Free tyrosine liberated due to β-casein degradation was measured as described earlier. Enzyme activity present per amount of enzyme is calculated as specific activity of the protease.
We have seen increasing protease activity in the initial substrate concentration range and then saturation of protease activity above concentration of 4.03 mg/ml β-casein (Figure 7). The graph as a result follows conventional Michaelis Menten kinetics. We calculated KM and Vmax from the corresponding double reciprocal plot i.e. Lineweaver Burk plot as shown in the inset graph (Table 2). KM is 5.47 mg/ml and Vmax is 588.23 μM/min.
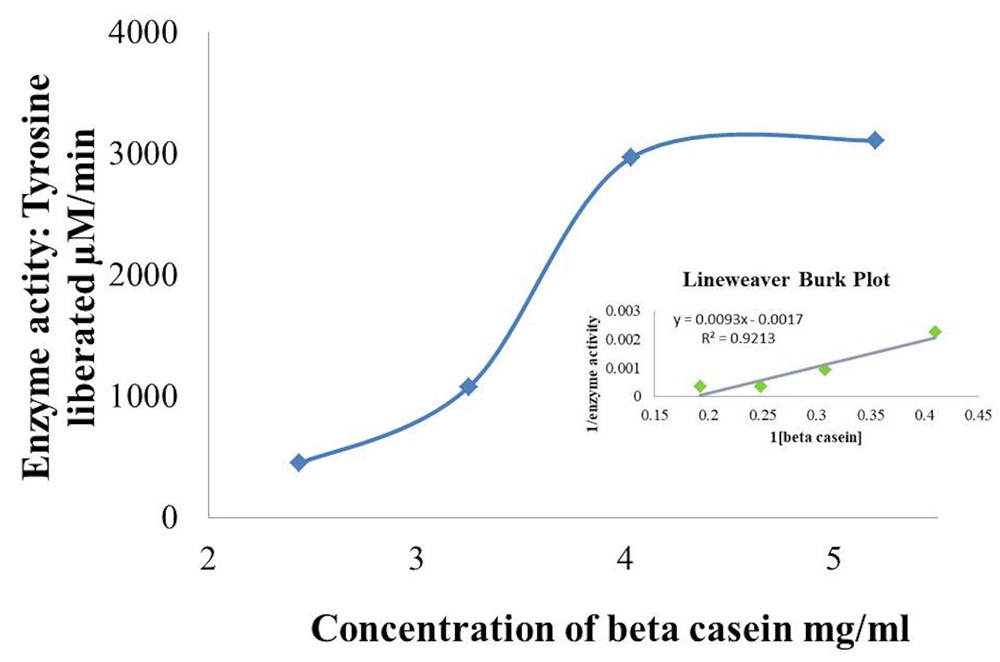
KM and Vmax obtained from the double reciprocal plot are 5.47 mg/ml and 588.23 µM/min respectively.
Our study concludes that mature leaves from Moringa oleifera contains a protease with an approximate molecular weight of 51kD, with an optimum temperature of 37°C and optimum pH of 8.0 for its caseinolytic property. This is the first report of purification of a protease from Moringa oleifera to our knowledge. Further determination of molecular characterization, substrate specificity and activity of the protease are required to determine its suitability for industrial applications.
Dataset 1: Enzyme kinetics data. Zip file containing underlying data of all enzyme activity assays with raw gel images 10.5256/f1000research.15642.d212249
We would like to acknowledge Mr Dipak Chandra Konar of Department of Chemistry, Bose Institute, for his help in using the protein purification set up, and spectrophotometric assays. Special thanks go to Prof. K. P Das, Bose Institute, for his support in this project.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
No source data required
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: DNA damage repair mechanism in plants under abiotic stress
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 30 Jul 18 |
read | read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)