Keywords
COVID-19; vaccine; seroconversion; efficacy; immunization.
This article is included in the Pathogens gateway.
COVID-19; vaccine; seroconversion; efficacy; immunization.
COVID-19 remains a global challenge.1 While the incidence of COVID-19 was reported to have decreased in recent months, recent reports have suggested that there has been a rising trend in COVID-19 incidence.2 This fluctuating incidence might be affected by the variants of concern,3 in which COVID-19 management also remained challenging,4 and the management of new variants of concern might differ from that of previous variants.5 The guidelines for COVID-19 management have been periodically published and updated.6 However, the efficacy of each treatment was inconclusive, particularly in the case of severe or critical illness.7–9 Therefore, the proper treatment of COVID-19 remains under investigation. While the potential of new drugs has been explored,10 the vaccination program appears to have the potential to end this pandemic.11
The COVID-19 vaccination program was introduced in early 2021 and implemented worldwide.12 This vaccination program was initially targeted to health workers, a population with high risk of infection, and continued to the public.12,13 To date, a wide variety of COVID-19 vaccines have been available, such as inactivated, messenger ribonucleic acid (mRNA), protein subunit, and vector vaccines.14 However, efficacy differs between vaccines, and the results from each study have varied.15 In this circumstance, conflict between pharmaceutical companies might occur. Therefore, the question of which vaccine has the best efficacy remains. In the content of vaccination, seroconversion was used to assess the early response of neutralizing antibody production.16 However, the report of seroconversion of COVID-19 vaccines varied in each study, particularly in the special cases with comorbidity.17–19 Moreover, to date, no study has directly compared the efficacy of COVID-19 vaccines. Therefore, the present study aimed to assess the indirect comparison of seroconversion rates among different COVID-19 vaccines using a network meta-analysis approach. Our present study provides preliminary evidence regarding potential COVID-19 vaccines.
A meta-analysis, following the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) protocols,20 was conducted to compare the seroconversion rates of different COVID-19 vaccine designs. To formulate a comprehensive comparison, the relevant articles were collected from PubMed, Embase, and Scopus, and the information of interest was extracted to compare the seroconversion rates among different COVID-19 vaccine designs.
Articles were included in our analysis if they met the following inclusion criteria: (1) assessed the seroconversion rate of a COVID-19 vaccine; and (2) provided standardized data to determine the seroconversion rate of a COVID-19 vaccine. The following articles were excluded: reviews, commentaries, letters to the editor, non-randomized controlled trials (RCTs), and double publications.
As of January 10, 2022, we searched for potential articles in PubMed, Scopus, and Web of Science. We determined potential COVID-19 vaccine designs to be involved in our study prior to searching the primary outcomes (seroconversion rate). We used the keywords adapted from medical subject headings: “COVID-19 vaccine” or “inactivatedCOVID-19 vaccine” or “mRNACOVID-19 vaccine” or “protein subunit COVID-19 vaccine” or “vector COVID-19 vaccine” and “efficacy” or “seroconversion”. The search was limited to RCTs and articles published in English. If a double publication was found, only articles with a larger sample size were included. We also browsed the reference list of relevant systematic reviews to obtain additional references. Subsequently, the following information of interest from the potential articles was extracted by two independent investigators: (1) first author name, (2) publication year, (3) study design, (4) age of patients, (5) sample size, (6) design of COVID-19 vaccine, (7) trade name of COVID-19 vaccine, (8) dosage of COVID-19 vaccine, (9) modified JADAD scale, and (10) seroconversion rate.
Prior to inclusion in our analysis, articles were appraised for quality using the modified JADAD scale. The scores ranged from 0 to 7. Scores of 5–7, 3–4, and 0–2 indicated high-, moderate-, and low-quality papers, respectively.21 Low-quality articles were excluded from the analysis. Using a pilot form, quality assessment was performed by two independent authors (JKF & MI). Discrepancies between the two authors were resolved by discussion.
The primary outcome was the seroconversion rate of the COVID-19 vaccine, defined as the level of geometric mean titer (GMT) of neutralizing antibodies of greater than or equal to four-fold from the baseline. The predictors were different COVID-19 vaccine designs. To identify the potential COVID-19 vaccine designs, an initial evaluation of the available data in PubMed, Scopus, and Web of Science was performed. Of those, inactivated, mRNA, protein subunit, and vector COVID-19 vaccines were available for the analysis.
Before analyzing the data, the potential publication bias and heterogeneity across the studies was assessed. Publication bias was assessed using an Egger test, and a p-value < 0.05 indicated a publication bias. Heterogeneity among the studies was evaluated using the Q test. A p-value < 0.10 suggested that heterogeneity existed among studies, and that a random effect model should be applied for the data analysis. Otherwise, a fixed-effect model should be used. The cumulative seroconversion rate of COVID-19 vaccines was determined using a single arm model meta-analysis by calculating the pooled seroconversion events from the total sample size. The effect size was presented using the seroconversion percentage and 95% confidence interval (CI). The data were analyzed using R package software (R package, MA, US, RRID:SCR_001905). The pooled seroconversion rates are summarized in a forest plot. The seroconversion rates between different designs of COVID-19 vaccines were compared by calculating the effect size of each COVID-19 vaccine design. The highest seroconversion rate was considered the highest efficacy, and the Confidence in Network Meta-Analysis software version 1.9.1 (Bern, Switzerland, RRID:SCR_016488) was used to outline the network diagram of comparison among COVID-19 vaccines.
We collected 8,070 potential papers from the databases. Of these, 23 duplication papers were found, and 7,966 papers had irrelevant topics; therefore, those papers were excluded. Subsequently, 81 papers were included for further full-text reviews. Among these, 19 reviews and 32 papers with insufficient data and were excluded. Finally, a total of 31 papers were analyzed to compare the seroconversion rates among COVID-19 vaccines.22–52 The process of article selection in our study is presented in Figure 1, and the characteristics of the papers included in our analysis are summarized in Table 1.
In the follow-up period of ≤ 15 days, 24 papers assessing the seroconversion rate of COVID-19 vaccines were collected. In total, the seroconversion rate was 91.1% (Figure 2A). The seroconversion rates of inactivated (Figure 2B), mRNA (Figure 2C), protein subunit (Figure 2D), and vector COVD-19 vaccines (Figure 2E) were 93.2%, 93.9%, 65.3%, and 54.7%, respectively. Subsequently, in the follow-up period of 16–30 days, the cumulative seroconversion rate of COVID-19 vaccines was 94.1% (Figure 3A). The seroconversion rates of inactivated (Figure 3B), mRNA (Figure 3C), protein subunit (Figure 3D), and vector COVID-19 vaccines (Figure 3E) were 96.0%, 94.8%, 91.2%, and 89.7%, respectively. Alternatively, in the follow-up period of 31–60 days, the pooled seroconversion rate of COVID-19 vaccines was 98.0% (Figure 4A). The seroconversion rate of inactivated (Figure 4B), mRNA (Figure 4C), protein subunit (Figure 4D), and vector COVID-19 vaccines (Figure 4E) were 98.5%, 98.6%, 98.5%, and 96.2%, respectively.
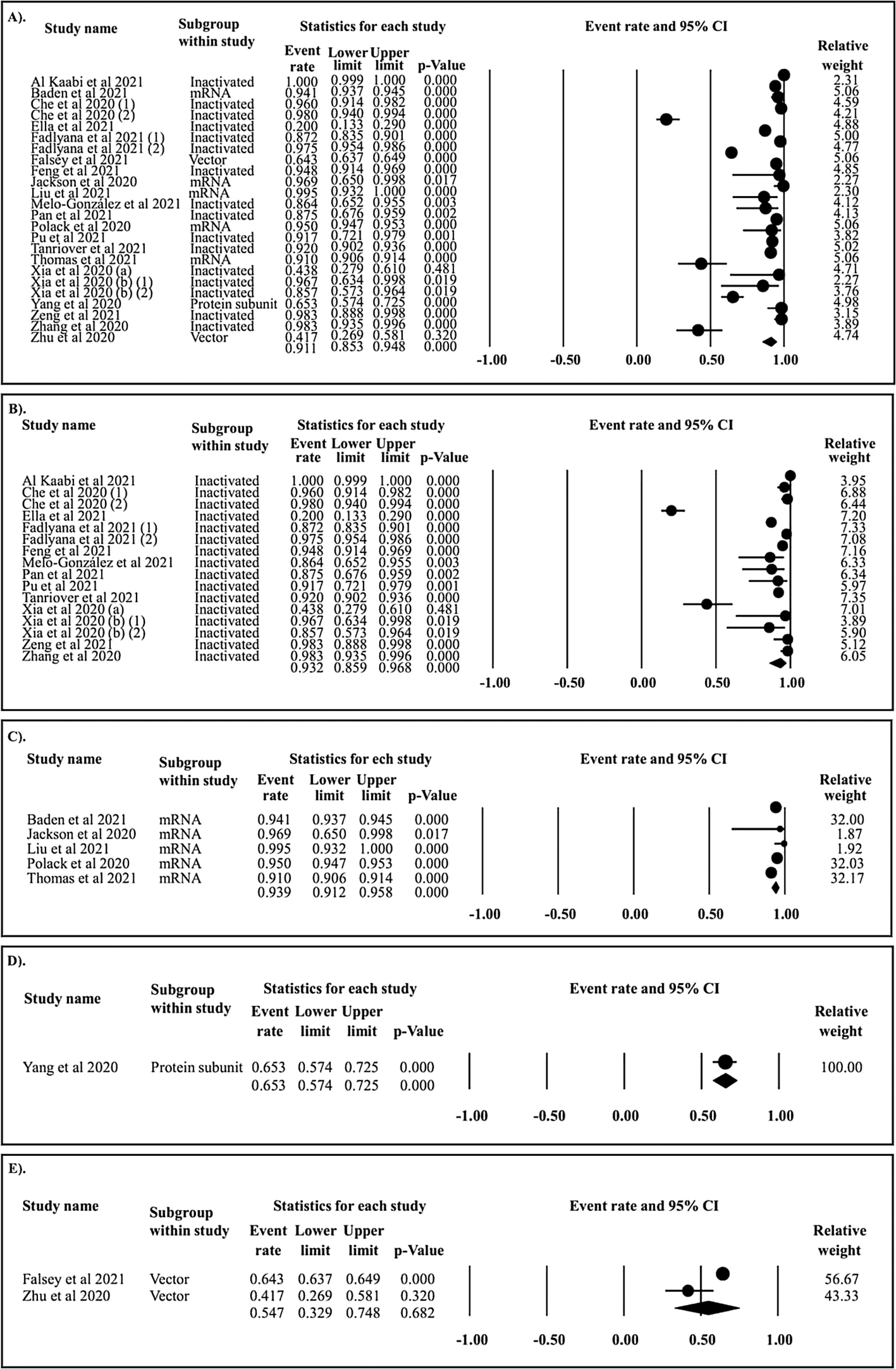
A). All vaccine types; B). Inactivated vaccine; C). mRNA vaccine; D). Protein subunit vaccine; and E). Vector vaccine.
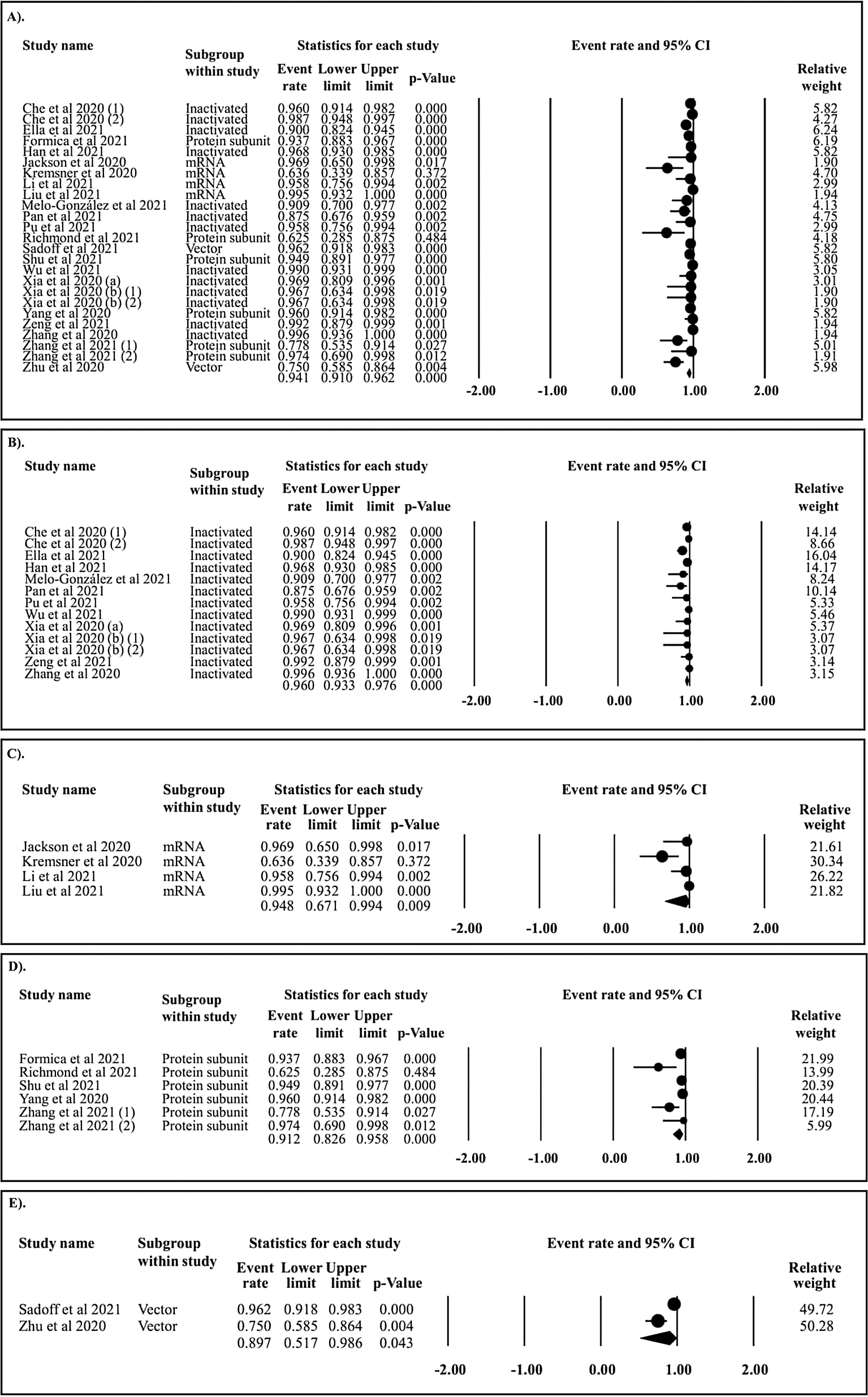
A). All vaccine types; B). Inactivated vaccine; C). mRNA vaccine; D). Protein subunit vaccine; and E). Vector vaccine.
In the follow-up period of ≤ 15 days (Figure 5A), the seroconversion rates of inactivated and mRNA COVID-19 vaccines were superior to those of the protein subunit and vector COVID-19 vaccines. In the follow-up period of 16–30 days (Figure 5B), the seroconversion rates of inactivated and mRNA COVID-19 vaccines were significantly higher than those of protein subunit and vector COVID-19 vaccines. In the follow-up period of 31–60 days (Figure 5C), the seroconversion rate of the vector COVID-19 vaccine was inferior to that of the inactivated, mRNA, and protein subunit COVID-19 vaccines. The network diagrams of seroconversion rates among different COVID-19 vaccine designs are presented in Figure 5D, 5E, and 5F for the follow-up periods of ≤ 15, 16–30, and 31–60 days, respectively. The summary of indirect comparison of seroconversion rate among COVID-19 vaccines is outlined in Table 2.
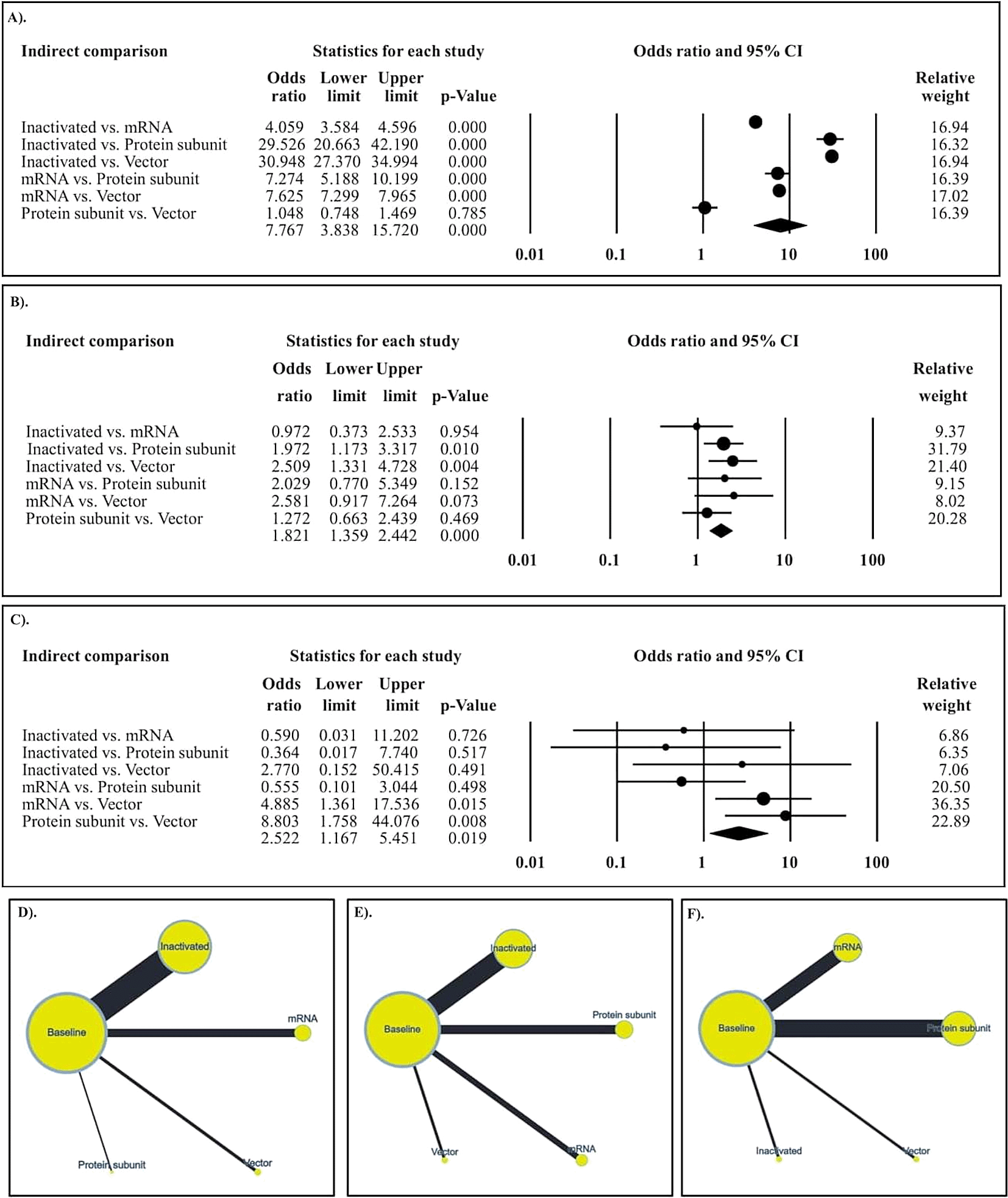
A). Follow up period at day ≤15; B). Follow up period at day 16-30; C). Follow up period at day 31-60; D). The networking among studies at day ≤15; E). The networking among studies at day 16-30; and F). The networking among studies at day 31-60.
In the follow-up period of ≤ 15 days, evidence of heterogeneity (p heterogeneity > 0.05) was observed in the models of analyses for all COVID-19 vaccines, inactivated COVID-19 vaccines, mRNA COVID-19 vaccines, and vector COVID-19 vaccines. Therefore, a random effect model was used. In the follow-up period of 16–30, evidence of heterogeneity was observed in all models of analyses; therefore, a random effect model was used. In the follow-up period of 31–60 days, all analyses were performed using a fixed effect model, because no evidence of heterogeneity was found (Supplementary files).20
Our study reported on the seroconversion rate among different COVID-19 vaccines in the follow-up periods of ≤ 15, 16–30, and 31–60 days. We revealed that, in the follow-up period of ≤ 15 days, the highest seroconversion rate was that of the inactivated vaccine. In the follow-up period of 16–30 days, the highest seroconversion rates were those of the inactivated and mRNA vaccines. Conversely, in the follow-up period of 31–60 days, the highest seroconversion rates were those of the inactivated, mRNA, and protein subunit vaccines. To date, our study is the first to report on the seroconversion rates among different COVID-19 vaccines; therefore, comparisons among meta-analyses were not discussed. However, similar meta-analyses have been performed to assess the efficacy of COVID-19 vaccines. In total, nine meta-analysis studies have been conducted.53–61 While the majority of these studies focused on side effects of the vaccine, they also reported the efficacy of COVID-19 vaccines by assessing the reduced risk of infection, mortality, hospitalization, and admission to the ICU. The findings revealed that the mRNA vaccine had the highest efficacy in preventing COVID-19 infection.59 Those previous meta-analyses support our findings that, besides the mRNA vaccine, the inactivated vaccine had a higher seroconversion rate than that of the protein subunit and vector COVID-19 vaccines.
The theory underlying the comparison of the efficacy of COVID-19 vaccines remains debatable. It has been widely proposed that various vaccines have various immunogens and may be engulfed, processed, and presented by antigen-presenting cells (APCs) along with MCH antigens to CD4+ T cells. Resultingly, cytokine synthesis may occur and may activate humoral and cellular responses, including antibody production, CD8+ T cell activation, and macrophage stimulation. Subsequently, B lymphocytes may differentiate into plasma cells and produce specific antibodies to protect against the infection.62,63 In the inactivated vaccine, the immunogenic property is a killed or modified virus containing the whole pathogen, virus fragments, or virus epitope.64 In the mRNA vaccine, the mRNA will be taken up by APC and translated into protein in situ. The mRNA encodes the full-length, pre-fusion stabilized spike protein (S) of SARS-CoV-2.14 In the protein subunit vaccine, the immunogenic property is specific isolated proteins from SARS-COV2 virus (S glycoprotein), which is responsible for receptor binding to cellular ACE-2.65–67 This type of vaccine is similar to a vector vaccine, in which the S protein is produced to confer protection against COVID-19.68 Of those possible mechanisms, the inactivated vaccine may target a wide variety of epitopes,69 and therefore, this type of vaccine may have a wide protection against COVID-19 variants of concern compared to other types of COVID-19 vaccines. However, vaccine specificity may differ, which requires evaluation by comparing the total GMT levels.
To the best of our knowledge, the present study is the first to report a comparison of seroconversion rates among different COVID-19 vaccine designs. Our study provided valuable evidence that inactivated and mRNA vaccines provided an early seroconversion rate, and the protein subunit vaccine achieved a similar seroconversion rate to that of inactivated and mRNA vaccines in the follow-up period of 31–60 days. The results of our research may be used as a basis for evaluating the development of COVID-19 vaccines in the future.13 We hope that future vaccine development may take into account the results of seroconversion rates between different vaccine designs, as we reported in our study. Therefore, the expected protective effects of the vaccine could be achieved. However, our present study only assessed the seroconversion rate in a short follow-up period. Further studies assessing a long-term follow-up period are required.
Our current study had several important limitations. First, we did not include potential confounding factors in assessing the efficacy of COVID-19 vaccines, such as comorbidity, nutritional status, and transmission area. Therefore, the probability of the dependent effect remains open to discussion. Second, the different report formats among studies required manual calculation for the precise seroconversion rate information. Therefore, there is the possibility of human error in interpreting seroconversion rates. Third, the vaccine and booster dosages varied among the studies. Therefore, false-positive findings might exist. Fourth, we only focused on the seroconversion rate, in which this evaluation was only effective in assessing the sensitivity of vaccines. Further studies evaluating cumulative GMTs might be required. Fifth, in the current study, vaccine safety was not evaluated. Therefore, further studies assessing safety are needed.
Our study revealed that the inactivated and mRNA COVID-19 vaccines provided the highest seroconversion rates at early follow-up. The protein subunit COVID-19 vaccine achieved a seroconversion rate similar to that of the inactivated and mRNA vaccines at the follow-up period of 31–60 days. Our study might contribute to better insight into the seroconversion rates of different COVID-19 vaccines.
All data underlying the results are available as part of the article and no additional sources of data are required.
Figshare. PRISMA checklist. DOI: https://doi.org/10.6084/m9.figshare.19236714.v220
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Idea/concept: GS, JKF. Design: GS, JKF, LW. Control/supervision: GS, LW, MA, KD, HH. Data collection/processing: MI, AA, HI, AAA, SL, US, TDY, EDN, FR, NR, RT, MVK, SS, MCH, UA, NH, NBF, VCL, UMP, FT, DAK, AIM, AP, EAP. Extraction/Analysis/interpretation: JKF, MI. Literature review: GS, JKF, MI. Writing the article: GS, JKF, MI. Critical review: GS, LW, MA, KD, HH. All authors have critically reviewed and approved the final draft and are responsible for the content and similarity index of the manuscript.
We thank Lembaga Pengelola Dana Pendidikan (LPDP) Republic of Indonesia for supporting this project.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Are the rationale for, and objectives of, the Systematic Review clearly stated?
Yes
Are sufficient details of the methods and analysis provided to allow replication by others?
Partly
Is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are the conclusions drawn adequately supported by the results presented in the review?
Yes
If this is a Living Systematic Review, is the ‘living’ method appropriate and is the search schedule clearly defined and justified? (‘Living Systematic Review’ or a variation of this term should be included in the title.)
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Immunology; Infectious diseases
Are the rationale for, and objectives of, the Systematic Review clearly stated?
Partly
Are sufficient details of the methods and analysis provided to allow replication by others?
Partly
Is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are the conclusions drawn adequately supported by the results presented in the review?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Biophysics; Thermodynamics; Kinetics; Immunology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 10 Mar 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)