Keywords
antibiotic resistance, enterobacterales, escherichia coli, street food, food microbiology
This article is included in the Antimicrobial Resistance collection.
antibiotic resistance, enterobacterales, escherichia coli, street food, food microbiology
This data contributes information about the antibiotic resistance profiles of Enterobacterales strains isolated from street food that will facilitate pathogen surveillance in Ecuador and Latin America. This data is useful for the scientific community to determine the presence of pathogenic Escherichia coli isolates and antibiotic resistance genes, including mobile colistin resistance genes, carbapenemases, quinolone resistance genes, and extended-spectrum β-lactamases present on Enterobacterales strains isolated from street food. Researchers and policymakers involved with the work related to the One Health initiative could also benefit from this data for retrospective and comparative analysis or epidemiological surveillance projects.1,2
Ready-to-eat street food was obtained in the streets of the city of Ambato, Ecuador, and processed the same day. A sharp sterile blade was used to cut the samples on sterile surfaces. 10 g of each sample was placed in sterile brain heart infusion broth (BHIB) (Merck, Darmstadt, Germany) in 90 ml, shaken on a rotator for 8-10 min, and incubated for 24 h at 37 °C. A large amount of broth was inoculated on MacConkey agar plates (Merck, Darmstadt, Germany), Cromocult Coliforms Agar (Merck KGaA, Darmstadt, Germany), and CHROMagar mSuperCARBA were incubated overnight at 37 °C under aerobic conditions. Further purification was performed on Macconkey agar.
The isolates were amplified by polymerase chain reaction (PCR), analysed using agarose gel electrophoresis and visualised with Sybr Safe DNA Gel Stain.3 For the identification of the isolated Enterobacterales, biochemical tests such as catalase, oxidase, TSI agar, Simmons citrate, lactose test, indole production, urea agar, methyl red test, and Voges-Proskauer were carried out and their interpretation was performed based on Bergey's manual.4 Additionally, the software for Automated Biometric Identification Systems (ABIS) was used to confirm the biochemical identification results.
Agar disk diffusion assays (Thermo Scientific Oxoid and Bioanalyse) on Mueller-Hinton Agar (Thermo Scientific Oxoid) were performed. Antibiograms tests were based on the measured diameter of the zones of inhibition and interpreted as sensible, intermediate or resistant by referring to CLSI breakpoints.5
The PCR test was performed according to the standardized protocol of the UTA RAM One Health research group6,7: 2.5 μL of DNA from each sample and 22.5 μL of PCR mix containing 12.5 μL DreamTaq PCR Master Mix (ThermoFisher Scientific, USA), 9 μL Nuclease-free water, 0.5 μL Primer 1 and 0.5 μL Primer 2 (final concentration of primers: 0.5 μM) were mixed to run PCR. The PCR conditions are reported in Supplementary Table S4. PCR products were analyzed by 1.2% agarose gel electrophoresis stained by Sybr Safe DNA Gel Stain (ThermoFisher Scientific, USA).
Hierarchical clustering
Hierarchical clustering was performed using the Euclidean correlation method and clustered by affinity.2 The MeV Multiexperiment Viewer software version 4.8.1 was used in this study.
The data presented show the frequency of isolation of Enterobacterales in 151 samples of ready-to-eat street food in Ambato, Ecuador (Figure 1). The specific characteristics (date of sampling, type of street food, location) of the samples were reported in Supplementary Table S1. A total of 145 isolates were analyzed, and the results of the biochemical tests were reported in Supplementary Table S2. Among them, 86 isolates corresponded to E. coli and 59 isolates to other Enterobacterales.
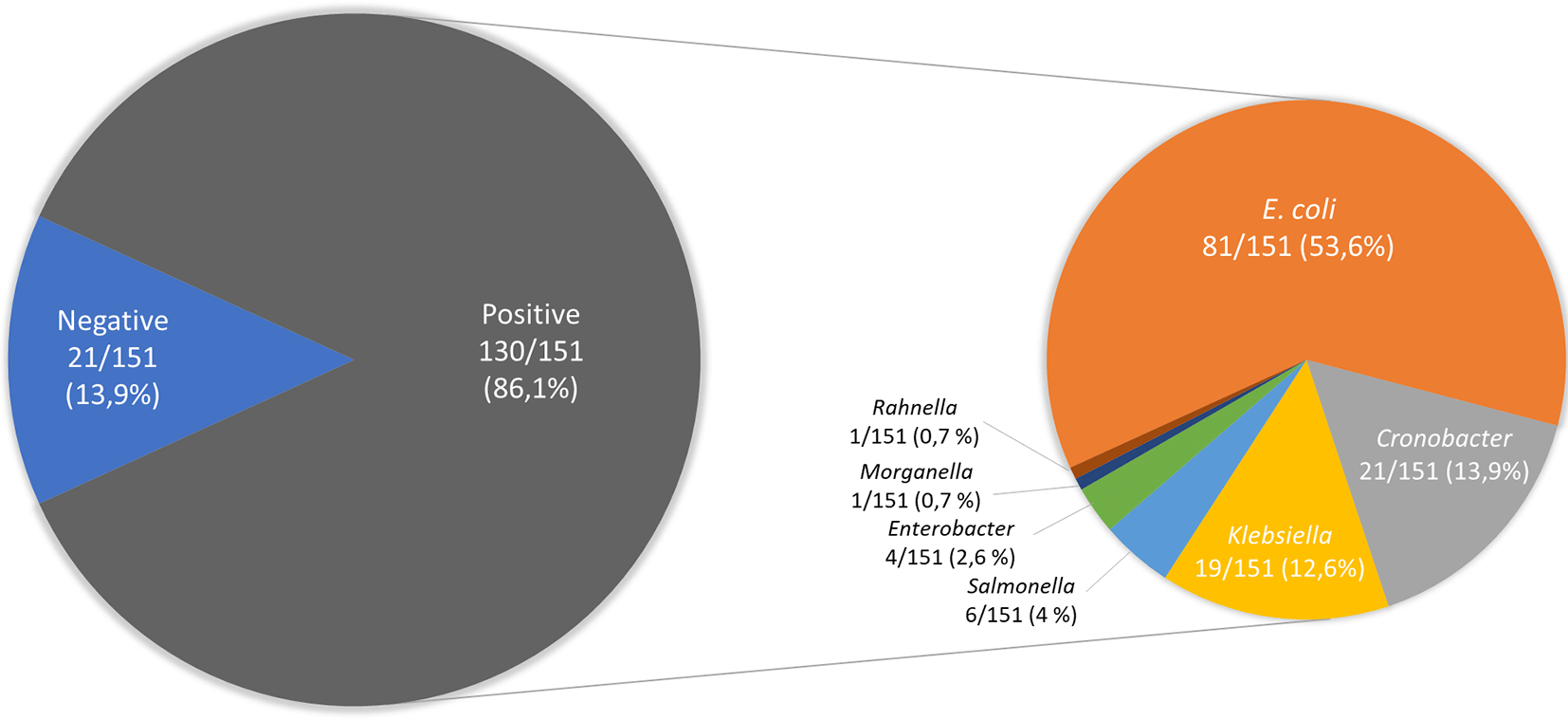
To visualize the relative similarity of the antimicrobial resistance patterns of the isolates, a hierarchical cluster analysis was performed using the results of the antibiograms, where the phenotypes ‘resistant’, ‘intermediate’, and ‘susceptible’ were observed as red, white, and blue colors respectively. Dendrograms and clustered data were assembled using the complete linkage method through Pearson correlation and sample leaf organization.7 For this purpose, the MeV Multiexperiment Viewer software version 4.8.1 was used.8 Figures 2 and 3 represent the resistance profiles and the hierarchical clustering of E. coli and the rest of Enterobacterales, respectively. The complete information is shown in Supplementary Table S3.

Red: resistant, White: intermediate, Blue: sensitive.
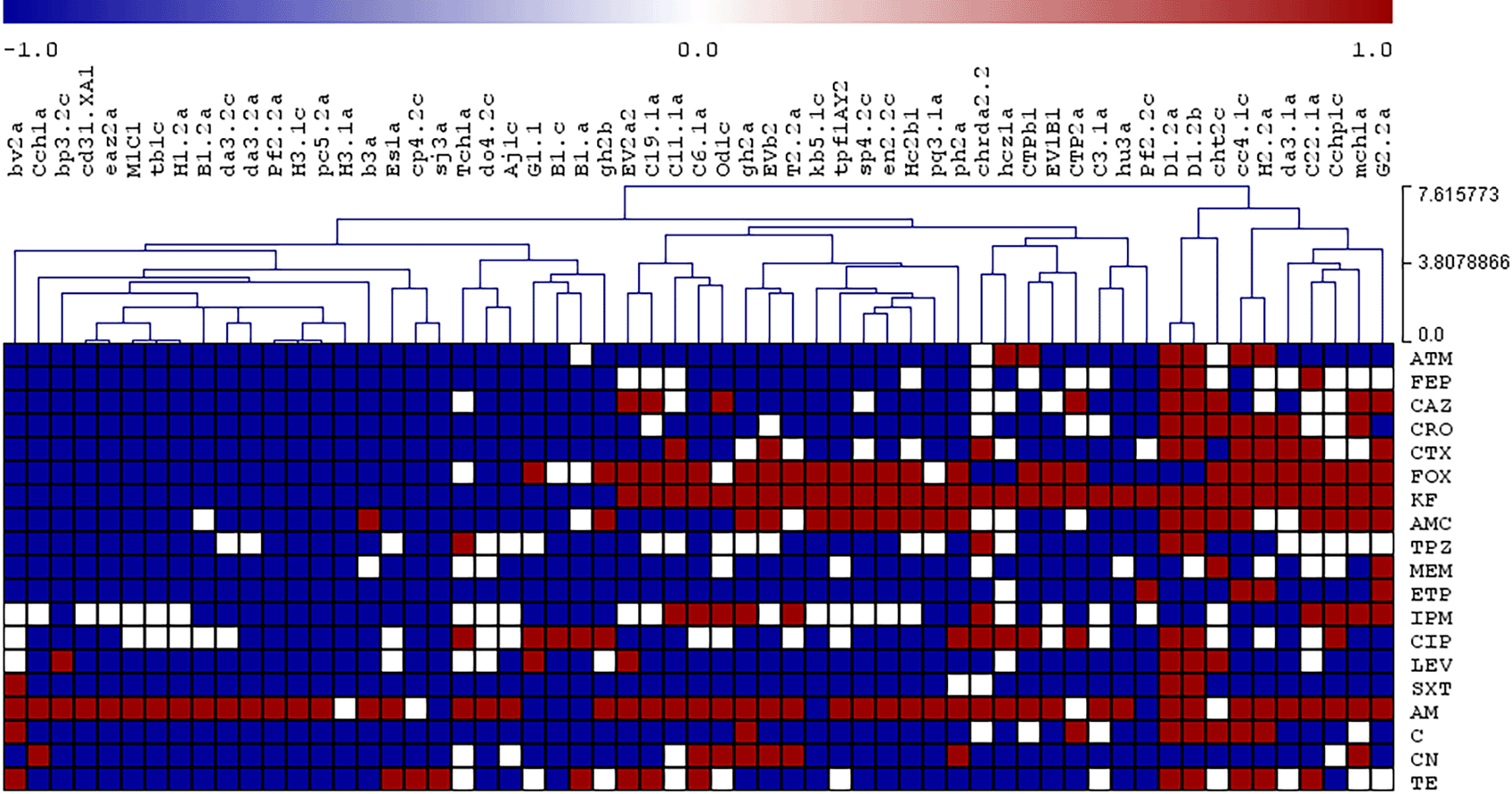
Red: resistant, White: intermediate, Blue: sensitive.
Abbreviations: TE: Tetracycline 30 μg, AM: Ampicilin 10 μg, KF: cephalotin 30 μg, C: chloramphenicol 30 μg, CIP: Ciprofloxacin 5 μg, CTX: Cefatoxime 30 μg, LEV: Levofloxacin 5 μg, FOX: Cefoxitin 30 μg, STX: Trimethoprim/sulphamethoxazole 25 μg, AMC: Amoxicyllin/ClavulanicAcid 30 μg, CN: Gentamicin 10 μg, CRO: Ceftriaxione 30 μg, FEP: Cefepime 30 μg, ATM: Aztreonam 30 μg, IPM: Imipenem 10 μg, TPZ: Piperacillin/Tazobactam 110 μg, ETP: Ertapenem 10 μg, MEM: Meropenem 10 μg, CAZ: Ceftazidime 30 μg.
The presence of diarrheagenic E. coli pathotypes present in ready-to-eat food was assessed in this study through the analysis of virulence genes related to the pathotypes. Only one isolate (C2.1c) was positive for the eae gene, suggesting the potential presence of enteropathogenic E. coli (EPEC) or enterohemorrhagic E. coli (EHEC). The β-lactamase resistance genes of Enterobacterales isolated in this study are reported in Table 1. Mobile colistin resistance genes or quinolone resistance genes were not found in the Enterobacterales isolates. The complete information about virulence genes and antibiotic resistance genes are available in Supplementary Table S5. The information about primers and PCR conditions were shown in Supplementary Table S4. The gel electrophoresis images are available at Supplementary figure S6. The disk difussion assays figures were shown at Supplementary figure S7.
Figshare project: https://figshare.com/projects/Data_on_antibiograms_and_resistance_genes_of_Enterobacterales_isolated_from_Ready-to-eat_street_food_of_Ambato_Ecuador/137014
This collection contains the following underlying data:
Figure 1. Occurrence of Enterobacterales on 151 samples of ready-to-eat street foods in Ambato, Ecuador. figshare. Figure. https://doi.org/10.6084/m9.figshare.19579087.v19
Table 1. Beta-lactamase resistance genes of Enterobacterales isolated from ready-to-eat food. figshare. Dataset. https://doi.org/10.6084/m9.figshare.19579099.v110
Figure 2 and 3. Antibiotic resistance profiles and hierarchical trees of Enterobacterales isolated from ready-to-eat street food in Ambato, Ecuador. figshare. Dataset. https://doi.org/10.6084/m9.figshare.19579267.v111
This collection contains the following extended data:
Supplementary table S1. Characteristics (Sample type, date, treatment type, location, coordinates) of the ready-to-eat food samples. figshare. Dataset. https://doi.org/10.6084/m9.figshare.19579108.v112
Supplementary table S2. Biochemical tests performed on Enterobacterales isolates from Ready-to-eat Street Food in Ambato, Ecuador. figshare. Dataset. https://doi.org/10.6084/m9.figshare.19579177.v113
Supplementary table S3. Antibiogram of Enterobacterales isolated from ready-to-eat Street food of Ambato, Ecuador. figshare. Dataset. https://doi.org/10.6084/m9.figshare.19579189.v114
Supplementary table S4. Primers used in this study and PCR conditions. figshare. Dataset. https://doi.org/10.6084/m9.figshare.19579198.v115
Supplementary table S5. Antibiotic resistance genes and virulence genes harbored by Enterobacterales isolates from ready-to-eat street food in Ecuador. figshare. Dataset. https://doi.org/10.6084/m9.figshare.19579207.v116
Supplementary figure S6. PCR results (positive electrophoresis images). figshare. Figure. https://doi.org/10.6084/m9.figshare.19729618.v117
Supplementary figure S7. Disk diffusion assay images-Antibiotic resistance evaluation of Enterobacterales isolated from ready-to-eat street food of Ambato, Ecuador. figshare. Figure. https://doi.org/10.6084/m9.figshare.19729630.v118
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
The authors would like to thank to the Directorate of Postgraduate Studies of the Department of Food and Biotechnology Science and Engineering, Universidad Técnica de Ambato.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bacterial Antibiotic Resistance, Molecular Microbiology, Molecular Epidemiology, Medical Microbiology.
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Yes
References
1. Chinen I, Campos J, Dorji T, Pérez Gutiérrez E: PulseNet Latin America and the Caribbean Network: Present and Future.Foodborne Pathog Dis. 16 (7): 489-497 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: antibiotic resistance, microbiology, phages
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: water microbiology, food microbiology, microbial source tracking
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 17 Jun 22 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)