Keywords
Viral Hepatitis, Hepatitis B virus, Extra-hepatic Manifestation, Lymphotropism
This article is included in the Pathogens gateway.
Viral Hepatitis, Hepatitis B virus, Extra-hepatic Manifestation, Lymphotropism
Viruses are distinguished intracellular obligate microparasites that infect a diverse spectrum of hosts ranging from bacteria to human beings. Their defining features consist of a simplistic genomic organization and a structural folding arrangement that compacts the size of the virion. Nonetheless, they display variations in genomic composition, varied protein expression, function, and efficacious patterns of evolution.1 Viruses commandeer their host’s metabolic pathways to facilitate their survival and propagation. The host invasion entirely depends upon the viral-host compatibility at the molecular level. These specific host viral interactions are necessary for the virus to sustain and maintain itself within the host.2 However, the complexity of pathogenesis can be summarized by the virus’s exploitation of its host through intricate viral-host protein interactions, which have been illustrated with network visualization studies.
The hepatitis B virus (HBV) is a prototypic member of the Hepadnaviridae family and has become a global pathogen with humans as its main target host. Viral hepatitis is notorious for causing chronic forms of hepatitis B (CHB) in approximately 250 million people worldwide and possess a 15–40 percent lifelong threat of terminal liver disease that mainly occurs in the form of liver cirrhosis and hepatocellular carcinoma (HCC).3,4 The virus is distinguished by its behavior within the host, where it utilizes reverse transcriptase as a mode of replication, which relies on host genetic repair mechanisms for its reproductive success.3 The virus carries a genome of approximately 3.2–3.8 Kb. However, it can cause a plethora of clinical manifestations. Although the virus is hepatotropic, it has been found beyond the bounds of the liver.5
The viral genome contains four ORFs that code the genes required for each viral process. The transcription of the HBV genome is initiated by four promoters; the core promoter is responsible for transcribing a 3.5-Kb mRNA for the core proteins that form the viral capsid, antigenic viral proteins (HbcAg) and the hepatitis B polymerase, which is the only enzyme native to the virus.5 The preS1 promoter transcribes a 2.4-Kb mRNA for the large surface Hepatitis B viral protein (LHB), which is the major component of the viral envelope as well as a sub viral particle. The preS2 promoter is responsible for the 2.1-Kb mRNA for the middle (MHB) and small (SHB) surface hepatitis B viral proteins of the viral envelope, which are also sub viral particles.6 The X promoter controls the expression of a 0.7-Kb mRNA for the only viral regulatory protein that regulates viral sustainability the host.6 Mostly known for its hepatotropic characteristics, HBV has also been found to have lymphotrophic properties.
The presence of intracellular mini-chromosomes, also referred to as covalently closed circular DNA as well as pre-genomic viral RNA to reverse transcribe the full viral genome are key indicators viral replication within host cells.5,7 This mini-chromosome acts as a reservoir for HBV proteins and persists within host systems even after infection clearance. The viral replication mechanisms are highly prone to mutations owing to the error-prone reverse transcription of an RNA intermediate, thereby accumulating a pool of viral quasispecies or variants of the same strains.7
Viral host protein interactome analysis depicts the processes of the virus within the host system, demonstrating the survival and propagation mechanisms used by the virus. An interactome network highlights the proteins that are frequently present and upregulated expression, which are considered as viral hub proteins.8 Moreover, these hub proteins depend on how well connected they are to host regulatory mechanisms. Thus, these hub proteins are necessary for normal cellular homeostasis. However, they are also widely exploited during pathogenesis as well.
Recent research on HBV has reported the possibility of extra-hepatic manifestation of the virus.9 Additionally, the human immune system is being researched for its possible contribution in viral propagation and persistence. These reports not only suggest the viral particle resides within the host immune cells, but also manipulate them for the successful propagation of viral hepatitis either by down regulating or upregulating immune responses away from homeostasis.10 Therefore, this review is a comprehensive analysis of the network generated through data derived by the hybrid text mining approach. This network highlights the contribution of the molecules, genes, and proteins of human peripheral blood mononuclear cells that play roles in the pathogenesis of HBV. The reason to consider host peripheral blood mononuclear cells is because they contain small fractions of all the white blood cell types. This network has been constructed based on the hypothesis that HBV invades, replicates, and manipulates host PBMCs genes and proteins to propagate its viral infection.
Text mining using a deep curation-hybrid approach
The deep-curation method involves screening of the scientific literature by experts, whereas text-mining method extracts biological data and their relevant interactions through keyword-based search from publication data using predictive algorithms and computational software. Although text mining is time saving, it has its own set of limitations that restricts its ubiquitous use. For example, the representative statement from a research article,11 “RIG-I expression in peripheral blood mononuclear cells was higher in responder than non-responder CHB patients treated with IFN-α therapy.” indicates that RIG-1 expression was higher, which would be a positive interaction for text mining. However, according to the real context of the text, RIG-1 expression is lower in patients with CHB who did not receive the therapy.11 Hence, manual curation is a necessary aide for refining data sets derived from text mining methods.
Data retrieval
The data pertaining to the direct or indirect interaction evidence between the HBV proteins and the host PBMCs proteins and genes were derived from scientific publications. These interaction data were derived from publications available in various repositories such as NCBI (https://www.ncbi.nlm.nih.gov/), PubMed (https://pubmed.ncbi.nlm.nih.gov/) and Google Scholar (https://scholar.google.com/) using keyword searches. The initial keywords ‘Hepatitis B’ and ‘Human’ were used, which generated 88,113 research articles on epidemiology, clinical-trials, vaccinations, therapeutics that were beyond the scope of the hypothesis. Subsequently, the keywords ‘Hepatitis B’ and ‘lymphotropism’ were used and retrieved 360 research articles. However, the literature was vague, and the information deviated from the key area of research. Similar issues were observed with the keywords ‘Hepatitis B’ and ‘Extrahepatic Manifestation’, which generated 29 hits but limited concrete information pertaining to our hypothesis. Eventually, an exhaustive list of keywords was prepared that contained all the known HBV genes and proteins, all molecules of host PBMCs such as dendritic cells, HLA, and synonyms for PBMCs such as white blood cells, lymphocytes, and leukocytes (https://docs.google.com/document/d/1NnNmHggs18num01nPO8G9i7LS80hwL8j23Wgu1L8K6g/edit). The list included relational keywords to identify a correlation between the viral and host proteins during the text mining, including inhibit, replicate, upregulate, downregulate, significant, and higher. The keywords were extracted by searching through the research articles that pertained to HBV and human host protein in PubMed published in the past ten years. The keywords were matched across all the abstracts downloaded from PubMed using keywords ‘HBV’ and ‘Human’ using Python scripts (https://docs.google.com/document/d/1iqiHhTwxzTe3DA9F_G3Qq2qh8f6uU4VT/edit).
Eligibility criteria
We identified 22,065 abstracts from publications in the format of PubMed IDs for the literature extracted as protein 1 and protein 2, which could interchangeably contain host or viral proteins that were relational to the keywords associated with the literature. However, out of the 22,065 abstracts, we chose 210 that contained keywords including host PBMCs, peripheral blood mononuclear cells, or lymphocytes in the first column and virus-associated keywords in the second column. Out of these 210 abstracts, there were 60 that contained no correlation with the hypothesis or were directly related to the keywords under consideration. Therefore, we used deep curation to select 198 abstracts, where 150 were from Python scripts and 48 were manually collected from Google Scholar. The interactions derived from the 198 abstracts and full-length papers were used to construct a network using cell designer.12
The curation of 198 research articles (77 PDFs and 121 abstracts) identified 28 genes, 92 proteins, and 20 PBMC molecules implicated in HBV pathogenesis. The 20 immune cell types were found to be permissive for the viral penetration and replication as per literature. Generally, HBV particles are found only in liver cells; however, the presence of replicative forms of HBV in host immune cells led to the further elucidation of 28 genes and 92 proteins that interact with one or the other viral genes and proteins. The genes were either hyper-methylated or down regulated during pathogenesis. These genes were necessary for certain immune functions, and polymorphisms in these immune genes could that predispose the host to viral chronicity and persistence (https://docs.google.com/spreadsheets/d/1sX9XCkfpadD0GLt79qp1ZtAVSlZ8UTRg/edit#gid=2019819854).
Proteins and cytokines involved in pathogenesis are usually immune proteins that aid in the sensing and eradication of pathogens through immune responses such as inflammation, antigen presentation, and amelioration by immune cells. We identified 52 PBMC proteins and 22 cytokines shown to play a substantial role in establishing the infection. Additionally, 11 proteins and 7 cytokines were downregulated, while 45 proteins and 15 cytokines were upregulated during viral infection. A summary of the interaction patterns of various molecules are depicted as the pathways visualized in cell designer (Figure 1).
Hybrid curation generated diverse associations based on the involvement of genes, proteins, extrahepatic molecules, and molecules implicated in vertical transmission. This led to the generation of four modules that emphasized and implicated each interaction type. The first module summarizes evidence for the presence of the replicative form of HBV within the host’s extrahepatic immune cells. The second module highlights direct or indirect involvement of PBMC proteins. These proteins interact with the viral proteins summarized in this module. A brief of the innate and adaptive immune response molecules implicated in infection have been discussed here. The second module leads to the third module, which elaborates on the HBV infected PBMC molecules that facilitate vertical transmission. Neonates are susceptible to infection from their infected mother’s immune cells that contain HBV in any form. The module discusses the factors which facilitate the mother to child transmission, which further leads to the neonate’s susceptibility to the infection.
Overall, these modules illustrate different dimensions of the extra-hepatic manifestation of HBV in host immune cells. They also provide insight into the mechanisms through which HBV manipulates the host immune system for its own persistence. A concise overview of all the overall mechanisms of extrahepatic manifestation pathways utilized by the virus has been demonstrated in Figure 1 which has been constructed using cell designer.
First module: Lymphotropism of HBV
The liver hepatocytes provide a permissive environment for the replication and propagation of HBV infection, consistent with the virus’s hepatotropic behavior.7 The active infectious forms of viral presence within the host cell are represented by several infectious markers such as cccDNA, pgRNA, viral mRNAs, proteins, and subviral particles.13 Several studies have investigated if these can function as markers in PBMCs, which would illustrate the viral behavior within the host white blood cells. The Stoll-Beker group in 1997 tried to identify forms of infectious markers in samples of serum and PBMCs from infected patients to detect the occurrence of viral DNA, HbeAg, and HBsAg. The peripheral blood mononuclear cells were found to have multiple copies of viral episomal DNA as well as mRNA of L, S and X viral genes, amongst which the X mRNA was found in the highest titer.13 Similarly, another research group investigated all the molecules from PBMCs to estimate the behavior of HBV with each molecule and found that monocytes as well as B lymphocytes had the highest infectious particle titer rate.14 Subsequently, evidence of infectious particles was also found in NK cell, T cell-CD4+, and T cell-CD8+ cells.14 Together, these studies confirmed the lymphotropic patterns of HBV.
RT-PCR was conducted of the total HBV genome and HBV episomal-cccDNA in peripheral blood populations in host cells from patients infected with HBV. HBV DNA was detected in 12 HBsAg patient serum samples, as well as PBMCs with less < 1% DNA was found in a covalently closed configuration.15 A recent study reported that the upregulation of HBV replication amongst actively proliferating peripheral blood lymphocytes.16 The results confirmed that the concentration of the HBV DNA increased proportionally to PBMC proliferation on day 12 from six healthy volunteers. On day 12 the total number of PBMCs was estimated to be 13.61 times more than the initial number of PBMCs seeded. Additionally, the HBV RNA and HBsAg were observed in peripheral blood population using RT-PCT and in situ hybridization, respectively.16 Another study was conducted to investigate the dynamics of HBV replication in actively proliferating peripheral blood populations and confirmed that HBV could replicate in proliferating PBMCs.17 When the distribution of varied forms of the viral genome in the plasma and PBMCs of chronically infected patients was investigated, tDNA was detected amongst PBMC samples derived from patients; however, the episomal form of the HBV genome was not detected in any sample. Viral genome integration in PBMCs was also reported in a study conducted where HBV DNA isolated from the serum, liver, and PBMCs was confirmed in the host genome.17 Viral genome integration was also observed in four out of seven PBMC samples taken from patients with chronic HBV hepatitis, which demonstrated the early integration of the HBV genome during the viral infection in PBMCs.17,18
The HBV has an affinity for the exosomes and extracellular vesicles associated with PBMCs. A study conducted in 2017 that isolated exosomes from the serum of CHB patients in the immune tolerant phase observed that the exosomes of the infected patients did in fact contain viral replicative markers.19 Interestingly, HBV replicative markers diffused into NK cells through the exosomes, which resulted in NK-cell dysfunction that interfered in cell signaling pathways such as NF-κB and MAPK. These results added another dimension to white blood cells susceptibility to HBV infection and revealed an undermined role of HBV-infected exosomes during the innate immune responses elicited by chronic HBV infection.20 These findings were followed by another study in 2018 that confirmed the presence of partial and complete forms of the virion within the extracellular vesicles and exosomes. The results demonstrated that the HBV infected vesicles were taken up by monocytes, followed by the upregulation of the PD-L1 protein that in turn, led to downregulation of CD69 T-cell populations and T-cell exhaustion. Moreover, the uptake of extracellular vesicles was also seen in dendritic cells and macrophages.21 A concise depiction of all the molecules have been depicted in Figure 2. Moreover, the proof of the evidence has been listed in Table 1 along with their journals which show direct relation of the findings.
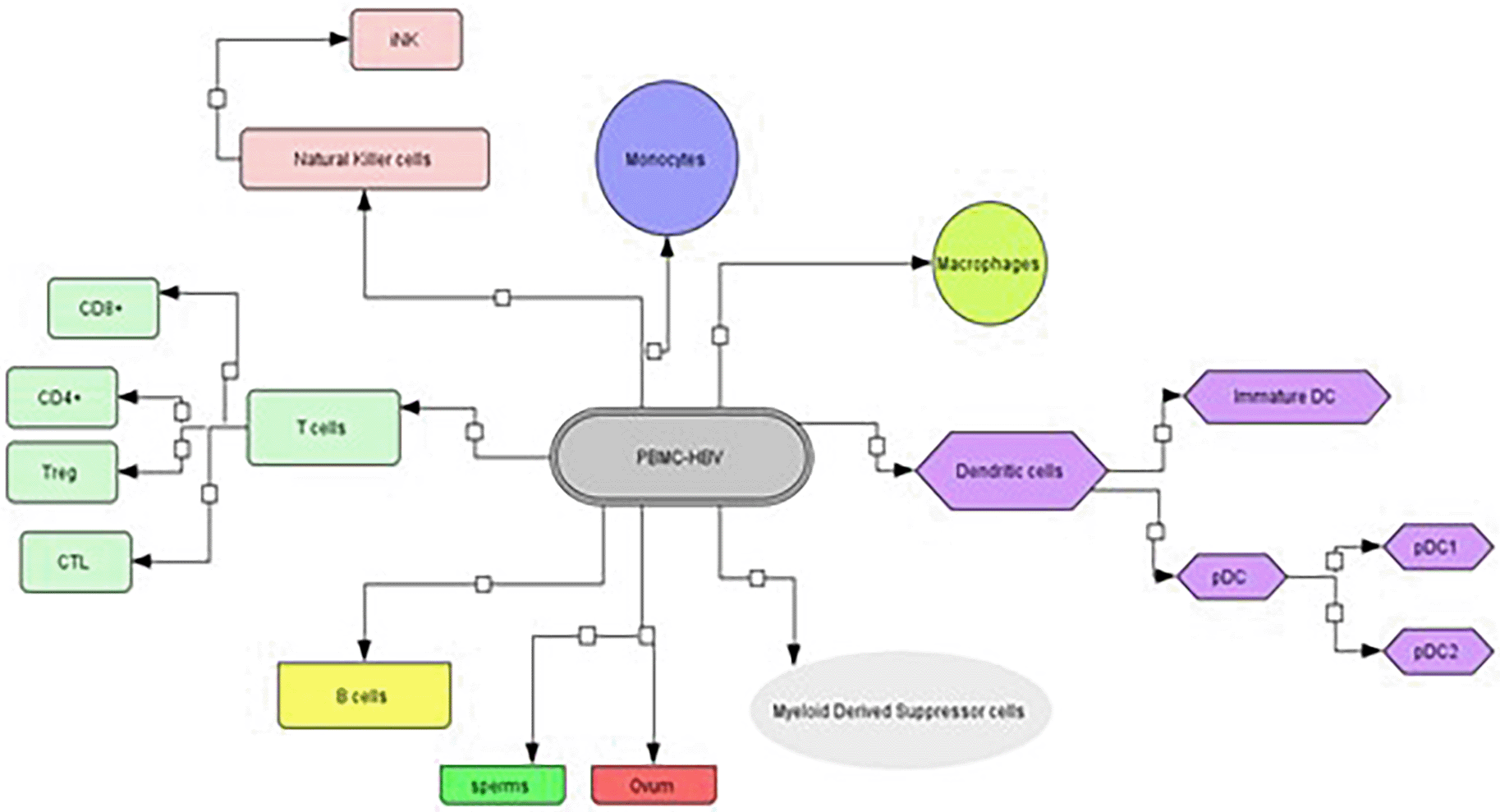
This is the demonstration of the diagrammatic view of the molecules of the white blood cell population where the HBV has been demonstrated to be found in its replicative forms. This network has been designed using cell designer.
These are the representations of the sections of the research articles which gives the proofs of the findings.
Second module: Role of host immune system proteins
This module highlights all the proteins in the PBMC population that have been experimentally found to interact with HBV during pathogenesis. Here, we summarize the interacting viral host proteins in a logical flow of events.
Most of the population that suffers from HBV infection fail to elicit any critical symptoms of the infection, yet nearly 15–40% of the cases develop into liver cirrhosis and hepatocellular carcinoma.10,13 Moreover, during infection, the inflicted population demonstrated elevated Alanine Aminotransferase or ALT levels. Along with increased alanine levels, high concentrations of viral DNA load were found in this population. The host defense system can suppress the infection for a prolonged duration without clinical manifestation of disease. Thus, the infected person even without clinical manifestation is a potential carrier of the disease.22 In contrast, chronic HBV infection during clinical manifestation inhibits effective immune responses against viral pathogenesis. This eventually culminates into an enduring challenge between the immune clearance response and the immune tolerance mechanism.23
The T-cell population is majorly responsible for clearance of HBV infection, especially the CD8+ population.24 The mechanism of T-cell impairment during the viral clearance response is still unknown. Various mechanisms have been hypothesized to explain the dysfunction of virus-specific T-cells. These mechanisms involve consistently high concentrations of the viral components and antigenic levels that impair the circulation and activity of T-cells.24 Alternatively, the high concentration of host factors such as IL-10 and TGF-β also interferes with the activity of T-cells by inducing T-cell exhaustion, which causes a loss of function in viral specific T-cells.25 T-cell exhaustion is associated with further elicitation of T-cell effector functions and proliferation inhibition. Exhausted T-cells can also induce an increased rate of apoptosis.26
There are various cytokines and regulatory molecules that are deregulated during infection and contribute to the progression of disease. In infected patients, Toll-like receptor (TLR) production is affected in monocytes. Ideally, activation of TLRs expressed by monocytes leads to the production of IL-10, IL-6, and TNF-α. TLRs have a critical function in the innate immunity that is required for inducing adaptive immune responses.27 An in vivo study showed that toll-like receptor 2 (TLR2) is necessary for eliciting effective virus-specific T-cell responses. The serum soluble TLR2 is required to downregulate viral mRNA in PBMCs.31 However, serum samples of CHB patients had lower concentrations of serum TLR2 compared to immune tolerant and inactive carriers. The PBMCs of CHB patients simultaneously expressed higher levels of IL-6 and TNF-α, which can cause liver injury. As with the increased concentration of the viral load during chronic infection, a few inhibitory receptors are overexpressed on the surface of exhausted T-cells.27 Receptors such as PD-1, CTLA-4, CD244 (2B4), and T-cell immunoglobulin domain and mucin domain 3 (TIM-3) are involved in inhibiting the function of T-cells expressing these receptors, which leads to an unresponsive state.28 During chronic HBV infection, there is an upregulation of TIM-3 in several species of immune cells, such as natural killer cells, dendritic cells, T-helper cells, cytotoxic T-lymphocytes, and macrophages. Tim-3 upregulation leads to impaired immune cell function. Moreover, elevated TIM-3 expression during infection inhibits host anti-viral immune responses, while deactivation of Tim-3 expression has a restorative effect. This highlights the relevance of the TIM-3 receptor during the HBV pathogenesis.28 Exhaustion can also be attributed to the inhibition of cellular telomerase enzyme production. It has been found that the expression of telomerase enzymes in PBMCs isolated from chronically infected individuals was severely impaired. This reduction in lymphocyte telomerase is associated with immunosenescence in the host cell population as well as the immunosuppressive condition in infected patients.29
Innate immune sensing proteins such as AIM2 (absent in melanoma 2) and IFI16 (interferon gamma inducible protein 16) are activated by various cellular stress inducing intracellular signals.30 These proteins subsequently activate caspase-1 (CASP1), which is an enzyme involved in the activation of inflammatory mediator molecules such as pro-IL-1β, which is converted into active secreted IL-1β. This pathway drives the host immune response towards a possible infection or disease state.30 During the infection, HBeAg or hepatitis B e-antigen inhibits the activation of the AIM2 and IFI16 inflammasomes that, during chronic infection, causes HBV mediated immune tolerance.31 Inhibition of IFI-16 also plays a role in the deactivation of the stimulator of interferon genes (STING)-dependent immune pathway. STING is an intrinsic DNA sensor that participates in the DNA sensor-STING-IFN-β pathway, which serves as an important mechanism against viral infections. It was reported that the IFI16 protein was expressed mainly in monocytes isolated from chronic hepatitis B patients but failed to elicit an effective response from the IFI16-STING pathway upon stimulation by its ligand VACV ds 70mer, and led to viral persistence.31
Text mining has highlighted few key studies that indicate a substantial role of the apoptotic pathway regulation by HBV during pathogenesis. Fas (apoptosis 1, CD95) and Fas ligand association facilitates deletion of T-cells through apoptotic pathways.32 The Fas ligand is mainly expressed on active T-cells that participate in TcR engagement. Activation of Fas by pathogenic antigens leads to up-regulation of Fas expression and sensitizes the mature T-cell to Fas mediate apoptosis. These (Fas) are mainly expressed on cytotoxic T-cells that are activated by HBV-related antigens on hepatocytes. Thus, this interaction between antigen-presenting cell hepatocytes, HBV antigens, and cytotoxic T-cells induces Fas-mediated apoptosis within the T-cells populations of patients with CHB.32 Similarly, the expression levels of the FAS, CASP3, CASP8 and CASP9 mRNA and proteins were compared in PBMC samples from healthy and infected individuals.33 The HBV infected group expressed higher levels of these apoptotic proteins than the healthy group, confirming HBV induced PBMC apoptosis is promoted when CASP8, CASP 9, and FAS are induced by both endogenous and exogenous pathways to activate CASP3, which acts as a common effector for these two pathways, resulting into viral-mediated cellular apoptosis.39 Another indirect apoptotic process PBMCs are susceptible to occurs through the upregulation of the programmed death-1 (PD-1) protein. PD-1 is upregulated on T-cells during CHB. The upregulation of PD-1 receptors on viral-specific CD8 T-cells populations is necessary to cause the T-cell dysfunction that results in high viraemia in patients with CHB.33 The role of natural killer (NK) cells is also associated with the upregulation and over expression of PD-1 during CHB infection. NK cells are also involved in chronic HBV infection. During the CHB, NK cells act as a rheostat for virus-specific T-lymphocytes. NK cells can elicit apoptosis in HBV-specific T-cells through the up-regulation of PD-1 expression.34
The asialoglycoprotein receptors (ASGPR) are C-type lectin receptors expressed mainly on mammalian hepatic cells.35 These receptors are actively involved in HBV infection in hepatocytes. Asialoglycoproteins present on hepatocytes interact with the preS1 viral protein that aids in the internalization of viral particles.36 Human immature dendritic cells express isoforms of asialoglycoprotein receptors that facilitate receptor-mediated endocytosis.37 However, the uptake of viral antigenic components by dendritic cells does not support the early steps of hepatitis B virus infection.38 On the contrary, Vyas et al. (2018) demonstrated that the placental expression of asialoglycoprotein receptors was implicated in HBV vertical transmission from mother to neonate. The upregulation of asialoglycoprotein receptor expression in the placenta was associated with hepatitis B surface antigen, indicating its role in vertical transmission HBV.39
Asialoglycoprotein receptors have also been found on various white blood cells. These are responsible for the formation of endocytic vesicles and its relevance in HBV infection was established in the third module as a component of vertical transmission of the virus. These receptors have also been associated with Hepatitis C virus infection in hepatocytes,40 as well as being expressed in immature dendritic cells and monocytes.41 However, the effect of these receptors during the HBV pathogenesis has not been described. Since the receptor isoforms are present on white blood cells, it is a potential target for the virus to enter the PBMC population and thus, it a viable target for therapeutic development.
Apoptotic pathway proteins have been identified in studies of HBV infected PBMCs. Apoptotic pathway modulation has been observed in hepatocytes and the HBx protein is involved in the modulation of these pathways.42 However, only a few proteins involved in apoptosis have been found in PBMCs. The proteins Fas, PD-1, and TIM-3 are overexpressed during the infection. The exact mechanism of TIM-3 is not understood in HBV PBMCs; however, in HIV cases, Tim-3 can recognize cells marked for apoptosis by the FG loop of the IgV domain that is crucial for the phagocytic clearance of apoptotic cells.42
Another very important aspect of viral initiation of apoptosis is apoptotic mimicry, which was hypothesised for HBV in 2003 by Peter Vanlandschoot and Geert Leroux-Roels (Vanlandschoot and Roels, 2003). This mechanism was also hypothesized for other viruses like HIV that use the apoptotic mimicry to enter the host cell and establish a successful infection.43 TIM-3 is one of the molecules hypothesized to be involved in initiating apoptotic mimicry. This pathway is one of the biggest revelations in HBV research and is highly underrated as this could serve as an important link of the entry of the viral particles in white blood cells, but it is still hypothesized.44
Third module: Vertical transmission
The third module is based on vertical transmission of HBV infection, which describes the transfer of HBV from infected mother to the neonate. The mother with HBV in any phase induces chronic forms of infection in her neonates. This module includes a discussion of this mode of transmission and the postulated mechanisms of the chronic infection and host immune cell behavior to decipher the global mechanisms HBV uses to manipulate the immune system.45
Maternal transmission of HBV infection to the fetus in the womb not only causes a persistent form of chronic infection in the fetus but also confirms the lymphoproliferative nature of the virus.46,47 The susceptibility of the chronic onset of infection amongst newborns was nearly ninety percent; this susceptibility of transmission significantly increased amongst mothers who tested positive for HbeAg antigen, whereas the neonates that test positive for HBsAg have the highest susceptibility for chronic infection, indicating the use of HbsAg as an indicator of infection.48
Along with the insights about transmission, it is necessary to characterize the factors that cause only chronic forms of HBV in neonates infected through vertical transmission. Therefore, it is important to understand the immunological mechanisms that cause the viral infection, including its unique infection characteristics and ability to trigger a non-classical response of innate immunity.49 Lack of response during infection is significantly characterized by the lack of stimulation of interferon type 1 genes during logarithmic expansion of the virus as well as pro-inflammatory chemokines and cytokines during the acute phase of the viral infection, which was confirmed by the in vivo and in vitro studies. It is still debated whether the virus suppresses the innate system or circumvents it altogether.50
To determine the mechanism to vertical transmission, a study reported that a maternal HBV infection in mice reduced the CD8+ T-cell immune response to the infection in progeny, thereby conferring HBV persistence.51 Further, it was found that a weakened T-cell immune response was due to live macrophages that were already susceptible to maternal precure antigen (HBeAg) that upregulated the otherwise inhibitor programmed death ligand 1 (PD-L1) and the polarization of macrophages upon repeated exposure.52
Asialoglycoproteins on hepatocytes were found to aid HBV in entering the host cell by eliciting endocytosis. However, asialoglycoproteins as well as their isoforms were found on the trophoblasts of the placenta of human pregnant infected mothers, as well as on the dendritic cells of infected mothers and the neonate.53 In a human population study conducted to isolate host factors that play a role in HBV vertical transmission, 12 HBsAg mothers along with their neonates were recruited for this study where asialoglycoproteins were found to be expressed highly during the infection. Therefore, it may be inferred that identification of the active role of asialoglycoprotein during HBV infection might provide an insight for a way for not only future therapies but may also be a mechanism of HBV extra hepatic manifestation.54
Persistence of CHB within neonates may be linked to an important defect in the response of viral-specific T-cells to infection that downregulates HBV-stimulated CD8+ and CD 4+ T-lymphocytes.55 Furthermore, infected woodchuck neonates were shown to have downregulated IFN-γ levels and a surge in TNF-α, which were identified as factors responsible for persistent CHB infection.56 This has also been observed in infected hosts who do not generate an antiviral immune response. Moreover, the exposure of hepatocytes to viral antigens in the absence of pro-inflammatory cytokines can lead to a dysfunctional T-cell immune response.57
Woodchuck models have been used to demonstrate that maternal HbeAg is a soluble protein that is able to cross the placental barrier and interact with fetal T-cell populations. It was found that HBeAg can mediate viral persistence by suppressing macrophage-dependent CTL responses. To further validate these results, woodchuck neonates were infected with a viral inoculum of a precure negative mutation that substantially reduced chronic disease symptoms.58
The infected maternal immune system can contribute to neonate immune tolerance. In a study of PBMC samples collected from 50 sets of mothers and their babies that were HBV positive, dendritic cells, T-cells, T-follicular helper cells, B-cells, cytokine profiles, and functional immune responses were analyzed using transcriptome profiling.59 The transcriptome analysis revealed that maternal immunological imprints associated with the disease were also found in neonates, with reduced levels of T-follicular cells and B-cells as well as lower induction of IL-21 levels observed. These parameters were linked to the persistence of CHB in neonates, providing a clear indication that neonates have lowered protective immunity due to the maternal infection.59
Recent reports of extra-hepatic manifestations have promoted a surge of research on the lymphotropic nature of the virus.60 A few studies have highlighted the presence of viral particles within host immune cells. HBV carrier PBMCs were linked to one of the methods of vertical transmission; in the study of a set of 312 HBV infected mothers, isolated PBMCs were used to investigate the pattern of their transmission from the mothers to the neonates.61 Seromarker analysis of neonate blood samples confirmed the transmission of viral DNA, viral antigen, and viral DNA-positive PBMCs. The existence of HBV-DNA-positive neonates was higher than the other serological markers tested. Despite these observations, few conclusions can be drawn regarding the exact mechanism of infection transmission through vertically. This is because there are several parameters that are still required to be assessed to understand how vertical transmission of HBV is initiated and regulated between host mother and neonate The vertical transmission of HBV through germ cells has been one of the leading yet under characterized factors of neonatal infection. Viral manifestation has been reported in ova of the infected individual; however, the integration of the HBV genome into the ova may alter the cell’s ability to yield a complete infectious virion.62 Indeed, a study with a sample size of 24 neonates of couples who were HBV positive showed that while 12 couples in which either adult had an infected germ cell either sperm or ova, none of their neonates manifested an infection.63,64 A concise depiction of all the molecules of module 2 and 3 have been depicted in Figure 3. Moreover, the proof of the evidence has been listed in Table 2 along with their journals which show direct relation of the findings to support our findings.
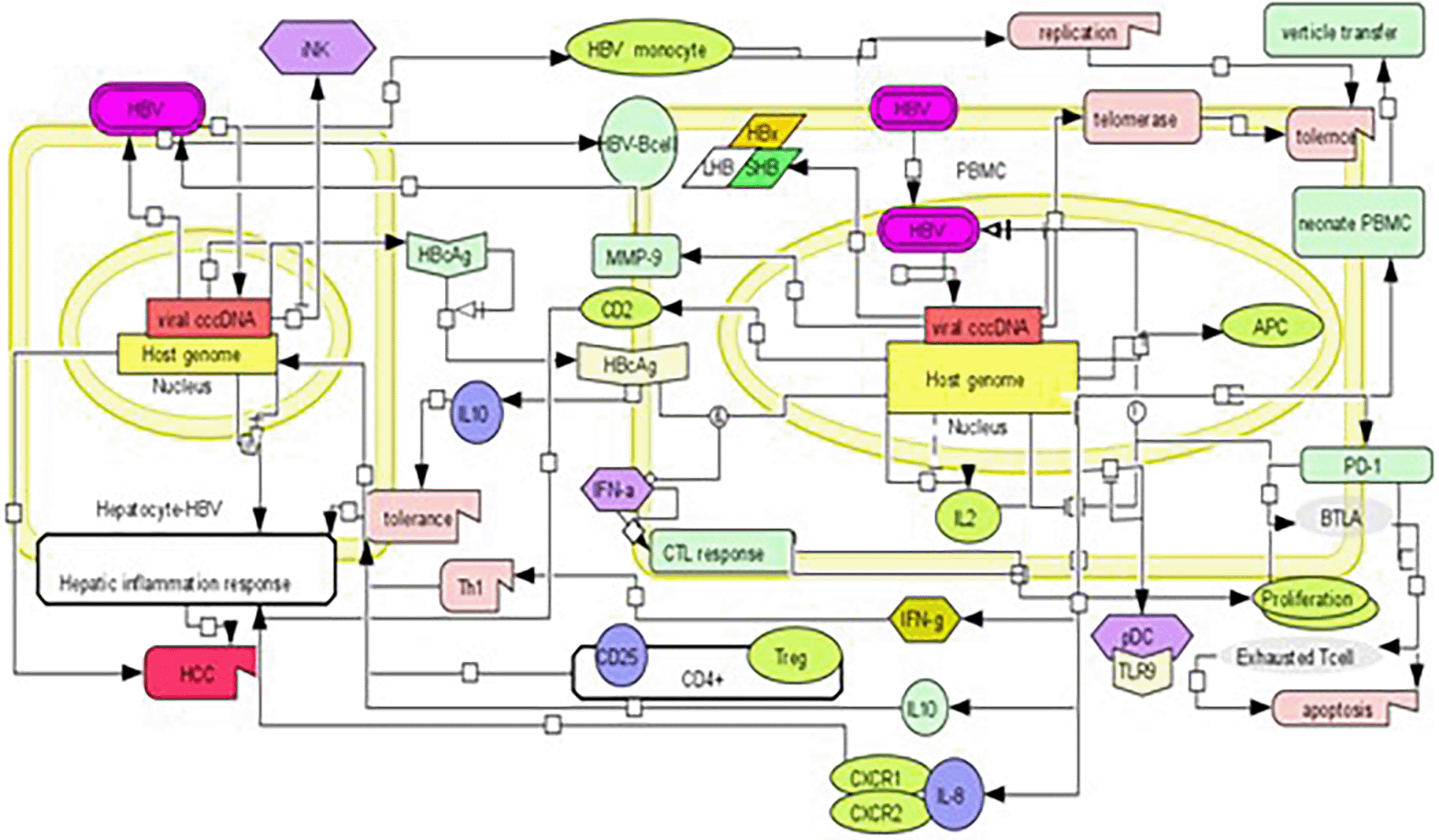
These proteins were identified by deep curation and are depicted in the form of a network constructed using Cell Designer.
These are the representations of the sections of the research articles which gives the proofs of the findings.
| Sno | Host cell/protein/gene | Viral Key gene/Protein | Statement of proof | Author | Journal | Year |
|---|---|---|---|---|---|---|
| 1 | IL-2 | HBsAg | PBMCs from HBV infected patients produced significantly lower levels of IL-2 than healthy controls. | Nagaraju et al. | Indian Journal of Gastroenterology | 1998 |
| 2 | sIL-2 | HBV | Serum sIL-2R levels correlate with the progression of liver damage in CHB patients. | Sawayama et al. | Digestive Diseases and Sciences | 1999 |
| 3 | IFN-γ | HBV | IFN-γ released by T-cells is critical for the anti-HBs clearance response. | Böcher et al. | Hepatology | 2000 |
| 4 | IL-6 | HBV | Elevated levels of IL-6 in serum occurs in PBMCs during HBV infection. | Song et al. | Chinese Journal of Hepatology | 2000 |
| 5 | IL-6 | HBV | IL-6 mediates early viral detection and controls during the initial progression of pathogenesis. | Hosel et al. | Hepatology | 2009 |
| 6 | IL-6 | HBV | Activated monocytes elicit IL-6 in the blood. IL-6 is associated with the progression of CHB and further HCC. | Lan et al. | Journal of Clinical and Translational Hepatology | 2015 |
| 7 | IFN-γ | HBV DNA | As IFN- γ increases, the quantity of HBV DNA is significantly reduced. | Dai et al. | Chinese Journal of Hepatology | 2001 |
| 8 | Telomerase | HBV | Decreased telomerase activity within PBMCs during HBV infection is attributed to immune dysfunction. | Fan et al. | Chinese Journal of Hepatology | 2000 |
| 9 | IL-18 | HBcAg | The severity of disease is proportional to the production of IL-18. The more severe the disease, the higher the IL-18 concentration. | Weng et al. | Molecular Cancer | 2012 |
| 10 | IL-12 | HBV | the DCs of patients with HBV have significantly reduced IL-2 stimulation and production compared with healthy individuals. | Wang et al. | World Journal of Gastroenterology | 2001 |
| 11 | PBMC-IL-2-infant | HBV DNA | PBMC-mediated IL-2 downregulation is related to HBV invasion in newborns and leads to HBV vaccine failure. | Wang et al. | Chinese Journal of Medical Genetics | 2002 |
| 12 | Membrane-interleukin 2 receptor/mIL-2R | HBV | mIL-2R expression on PBMC surfaces of patients with HBV are lower than the control group. | Wang et al. | World Journal of Gastroenterology | 2001 |
| 13 | IL-10 | HbcAg | IL-10 production is stimulated by HBcAg in PBMCs and may contribute to virus-derived immune tolerance. | Hyodo et al. | Clinical and Experimental Immunology | 2004 |
| 14 | hFGL2 | HBV | HBV-induced hFGL2 prothrombinase/fibroleukin expression initates CHB amongst viral exposed patients. | Fan et al. | Chinese Journal of Medical Genetics | 2004 |
| 15 | hFGL2 | HBc | HBc and HBx viral proteins enhance hFGL2 transcription via c-Ets-2-dependent signaling in MAPK pathways. | Han et al. | Journal of Biochemical Chemistry | 2008 |
| 16 | hFGL2 | HBx | HBc and HBx viral proteins enhance hFGL2 transcription via c-Ets-2-dependent signaling in MAPK pathways. | Han et al. | Journal of Biochemical Chemistry | 2008 |
| 17 | CD1a (DC) | HBV | On the surface of DCs, IL-12, HLA-DR, CD1a, CD80, and CD86 expression is reduced in HBV led HCC patients. | Wang et al. | Chinese Journal of Medical Genetics | 2005 |
| 18 | CD80 (DC) | HBV | On the surface of DCs, IL-12, HLA-DR, CD1a, CD80, and CD86 expression is reduced in HBV led HCC patients. | Wang et al. | Chinese Journal of Medical Genetics | 2002 |
| 19 | CD86 (DC) | HBV-HCC | On the surface of DCs, IL-12, HLA-DR, CD1a, CD80, and CD86 expression is reduced in HBV led HCC patients. | Wang et al. | Chinese Journal of Medical Genetics | 2005 |
| 20 | IL-12 (DC) | HBV-HCC | On the surface of DCs, IL-12, HLA-DR, CD1a, CD80, and CD86 expression is reduced in HBV led HCC patients. | Wang et al. | Chinese Journal of Medical Genetics | 2005 |
| 21 | IFN-α-5 | HBV | The IFNa-5 induction increases Th2-type responses in CHB and reduction of the Th1/Th2 ratio in CHB patients. | Ariyasu et al. | In vitro Cellular and Developemental Biology-Animal | 2005 |
| 22 | IFN-α-8 | HBV | IFNa-8 increases Th1/Th2 ratios in PBMC of CHB patients. | Toshio Ariyasu et al. | In vitro Cellular and Developemental Biology-Animal | 2005 |
| 23 | TNF/IFN-γ | HBV | HBV infection inhibits the production of INF-g and TNF-a in PBMCs, which inhibits immunosuppressive agents. | Tang et al. | World Journal of Gastroenterology | 2005 |
| 24 | CD4+ | HBV | CD4+/CD25+ T-regs help modulate immune effectors during HBV infection. | Xu et al. | Journal of Immunology | 2006 |
| 25 | CD25 | HBV | CD4+/CD25+ T-regs help modulate immune effectors during HBV infection. | Xu et al. | Journal of Immunology | 2006 |
| 26 | FOXp3 | HBV | Levels of FoxP3(+)-cell and inflammatory cell infiltration are significantly increased in HBV patients. | Xu et al. | Journal of Immunology | 2006 |
| 27 | CD58 | HBV | HBV pathogenesis leads to elevated CD58 levels in PBMCs derived from patients with hepatitis B. | Li Sheng et al. | World Journal of Gastroenterology | 2006 |
| 28 | PD-1 | HBV | HBV induces CD8+ T-cells and PD-1 upregulation, which causes T-cell dysfunction in CHB patients. | Peng et al. | Molecular Immunology | 2008 |
| 29 | CD-2 | HBV | CHB severity can increase CD2 levels and lead to tissue damage. | Li et al. | Cellular and Molecular Immunology | 2008 |
| 30 | IL-4 (infant) | HbcAg | Infant PBMCs positive for HBV DNA in utero corelates with high interleukin-4 transcription. | Xu et al. | chinese Journal of Pediatrics | 2009 |
| 31 | IL-17 | HBV | HBV pathology related to liver fibrosis is attributed to an increase in IL-17. | Xu et al. | Chinese Journal of Cellular and Molecular Immunology | 2009 |
| 32 | TLR9 | HBV | TLR9 expression in pDCs during CHB was significantly lower in comparison to healthy controls. | Xie et al. | Microbes and Infection | 2009 |
| 33 | TGFb1 | HBV | TGFb1 was detected in DC-primed CD4+ T-cell supernatants. | Hong et al. | Chinese Journal of Hepatology | 2009 |
| 34 | APOBEC3G | HBV | APOBEC3G expression and subcellular localization in peripheral blood mononuclear cells was observed in CHB patients. | Chen et al. | Chinese Journal of Hepatology | 2010 |
| 35 | Th17 | HBV | HBV progression was attributed to Th17 cells and caused liver injury by inducing IL-10. | Wu et al. | Journal of Gastroenterology and Hepatology | 2010 |
| 36 | NF-κB | HBV | During CHB, NF-κB expression in DC from patients with chronic severe hepatitis B was up-regulated. | Jian et al. | Chinese Journal of Hepatology | 2010 |
| 37 | IL-21 | HBV | IL-21 is upregulated during HBV and important for the development of severe liver inflammation. | Hu et al. | Journal of Viral Hepatitis | 2011 |
| 38 | CXCR1 | HBV | During HBV infection, CXCR1, CXCR2, and IL-8 protein expression was higher in high HBV titre groups than the low HBV titre group. | Bi et al. | Chinese Journal of Cellular and Molecular Immunology | 2012 |
| 39 | CXCR2 | HBV | During HBV infection, CXCR1, CXCR2, and IL-8 protein expression was higher in high HBV titre groups than the low HBV titre group. | Bi et al. | Chinese Journal of Cellular and Molecular Immunology | 2012 |
| 40 | CCR2 | HBV | During HCC caused by HBV, the receptors CXCR2, CCR2, and EP400 are present on patient PBMCs. | Shi et al. | European Journal of Cancer | 2014 |
| 41 | IL-8 | HBV | During HBV infection, CXCR1, CXCR2, and IL-8 protein expression was higher in high HBV titre groups than the low HBV titre group. | Bi et al. | Chinese Journal of Cellular and Molecular Immunology | 2012 |
| 42 | HBSP | HBV | A novel splice-generated protein (HBSP) was detected in human hosts with CHB and elicited immune responses. | Bayard et al. | Journal of Viral Hepatitis | 2012 |
| 43 | B- and T- lymphocyte attenuator (BTLA) | HBV | BTLA receptor upregulation leads to T-cell exhaustion by restricting T-cell responses to CHB. | Cai et al. | Journal of Gastroenterology | 2013 |
| 44 | CCR2 | HBV | In HCC caused by HBV, the CXCR2, CCR2, and EP400 receptors are present in patient PBMCs. | Shi et al. | European Journal of Cancer | 2014 |
| 45 | EP400 | HBV | In HCC caused by HBV, the CXCR2, CCR2, and EP400 receptors are present in patient PBMCs. | Shi et al. | European Journal of Cancer | 2015 |
| 46 | HMGB1 | HBV | During chronic and acute HBV, the high-mobility group box 1 (HMGB1) proteins are significantly up-regulated compared with the healthy control. | Li et al. | Journal of Viral Hepatitis | 2014 |
| 47 | ROR-γt | HBV | CHB-serum enhances retinoic acid-related orphan receptor-γt (RORγt) during HBV infection. | Li et al. | Journal of Viral Hepatitis | 2014 |
| 48 | TLR-4 | HBV | Enriched HMGB1 in CHB patients leads to liver injury and inflammation due to the shift from Treg/Th17 to Th17 dominance, which is regulated by the TLR4-IL-6 pathway. | Li et al. | Journal of Viral Hepatitis | 2014 |
| 49 | TLR-4 | HbeAg | HBeAg may elevate TLR3, TLR4, and PD-1 expression and inhibit lymphocytic proliferation as a mechanism of viral immune tolerance. | Chen et al. | Hepatology International | 2017 |
| 50 | sCD40L | HBV | In CHB patients, B-cells can be activated by sCD40L and function as APCs to induce HBV-specific cytotoxic T-lymphocytes. | Liu et al. | Antiviral Research | 2018 |
| 51 | MOV10 | HBV | In CHB patients, MOV10, A3G, and IFN-α expression was significantly lower than in the control group. | Song et al. | Journal of Microbiology | 2015 |
| 52 | A3G | HBV | In CHB patients, MOV10, A3G, and IFN-α expression was significantly lower than in the control group. | Song et al. | Journal of Microbiology | 2015 |
| 53 | IFN-α | HBV | In CHB patients, MOV10, A3G, and IFN-α expression was significantly lower than in the control group. | Song et al. | Journal of Microbiology | 2015 |
| 54 | Galectin-3 | HBV DNA | Peg-IFNa-2a might lead to inhibition of viral DNA replication by upregulating galectin-3 expression in PBMCs. | Li et al. | Chinese Journal of Hepatology | 2014 |
| 55 | GR-α | HBeAg, pre-S1Ag | GRα mRNA expression differed between CHB patients and healthy controls. | Mei et al. | Molecular Medicine Reports | 2015 |
| 56 | Moesin | HBV | Amongst infected patients twelve differentially expressed proteins including moesin and NADH dehydrogenase [ubiquinone] iron-sulfur protein 3 were involved in biological processes such as cell kinase signaling and transcription. | Zhang et al. | Chinese Jounal of Hepatology | 2014 |
| 57 | NADH dehydrogenase (ubiquinone) iron-sulfur protein 3 | HBV | Twelve differentially expressed proteins including moesin and NADH dehydrogenase [ubiquinone] iron-sulfur protein 3 were involved in biological processes such as cell kinase signaling and transcription. | Zhang et al. | Chinese Jounal of Hepatology | 2014 |
| 58 | TLR-2 | HBV | CHB patients had increased TLR2, TNF-α, and IL-6 expression in PBMCs. | Huang et al. | Antiviral Research | 2015 |
| 59 | Glutathione-S-transferase P1 (GSTP1) | HBV | Significantly higher GSTP1 methylation was observed in acute-to-chronic hepatitis B liver failure group of non-survivors than survivors. | Gao et al. | Alimentary Pharmacology and Therapeutics | 2017 |
| 60 | TLR-3 | HbeAg | HBeAg inhibits lymphocyte proliferation and is accompanied by increases in TLR3, TLR4, and PD-1 expression and reduced IFN-γ production. This mechanism may be responsible for immune tolerance. | Chen et al. | Hepatology International | 2017 |
| 61 | Fas (CD95) | HBV | Higher levels of apoptosis correlated with higher Fas expression on PBMC surfaces in CHB patients when compared with controls. | Bendary et al. | Archives of Virology | 2016 |
| 62 | STAT1 | HBV | During HBV infection, the expression of host molecules STAT1 and IRF9, and the antiviral protein MxA was considerably lower than in the control group. | Hao Zhang et al. | Clinical Laboratory | 2016 |
| 63 | IRF9 | HBV | Expression of host molecules (STAT1, IRF9) and antiviral protein (MxA) was considerably lower in the control group compared with patients with HBV. | Hao Zhang et al. | Clinical Laboratory | 2016 |
| 64 | MxA-antivr | HBV | Expression of host molecules (STAT1, IRF9) and antiviral protein (MxA) were considerably lower in the control group compared with patients with HBV. | Zhang et al. | Clinical Laboratory | 2016 |
| 65 | Rubicon protein | HBV | The Rubicon protein was high in peripheral blood mononuclear cells in patients with HBV infection significantly promoted HBV replication. | Wan et al. | Cellular and Molecular Immunology | 2017 |
| 66 | IFI16 | HBV | IFI16 correlated to the stage of inflammation in CHB patients and could contribute to the HBV pathogeneses-related liver damage. | Pang et al. | BMC Gastenterology | 2018 |
| 67 | RIG-1 | HBV | The expression of RIG-I in PBMCs was higher in responder than inert CHB patients that underwent IFN-α therapy, demonstrating an inverse relationship between RIG-I and HBV infection. | Wu et al. | Antiviral Therapy | 2018 |
| 68 | RIG 1 | HBV | RIG-I functions as an HBV recognition sensor that activates innate signaling. | Wu et al. | Antiviral Therapy | 2018 |
| 69 | KLRG1 | HBV | Increased KLRG1 expression in NK cells occurs during HBV and reduces the secretion of IFN-γ in NK cells. | Li et al. | Chinese Journal of Hepatology | 2018 |
| 70 | α-manossidase 1 | HBV | Upregulation of α-mannosidase I in PBMCs may be important for HBV immune escape. | Jiang et al. | Viral Immunology | 2016 |
| 71 | NKG2D | HBV | The frequency of NKG2D+ NK cells in PBMCs was significantly higher in CHB patients than in healthy controls. | Wang et al. | Scientific Report | 2017 |
| 72 | MVP | HBeAg/HBsAg | The MVP virus-activated protein is increased in PBMCs of CHB patients when compared to healthy individuals. | Han et al. | International Union of Biochemistry and Molecular Biology | 2019 |
| 73 | CD59/Protectin | Core | An HBV core protein expressed in PBMCs forms a complex with CD59 to downregulate and sensitizes hepatocytes, resulting liver damage. | Yu et al. | Viruses | 2019 |
| 74 | Ly6/uPAR | HBV | Promotes CDC to cause persistent liver inflammation. | Yu et al. | Viruses | 2019 |
| 75 | IL-1β | HBV polymerase | HBV causes significant reduction in IL-1β production by inhibiting inflammasome pathways in which HBV-Pol is an essential requirement. | Xu et al. | Cellular & Molecular Immunology | 2018 |
| 76 | AIM2 | HBeAg | HBeAg inhibits the inflammasomes AIM2 and, IFI16 in CHB, causing immunotolerance. | Chen et al. | Viral Immunology | 2018 |
| 77 | STING | HBV | IFI16, DDX41, MRE11, and STING are highly expressed in CHB patients. | Chen et al. | Virology | 2020 |
| 78 | DDX41 | HBV | IFI16, DDX41, MRE11, and STING are highly expressed in CHB patients. | Chen et al. | Virology | 2020 |
| 79 | MRE11 | HBV | IFI16, DDX41, MRE11, and STING are highly expressed in CHB patients. | Chen et al. | Virology | 2020 |
| 80 | cGAS | HBV | HBV passively escapes detection by viral DNAs through the cGAS/STING pathway | Lauterbach et al. | Virology | 2020 |
| 81 | GP73 | HBsAg | HBsAg is necessary for the activation of GP73 expression and permits HBV replication in cell lines such as HepG2 and Huh7. | Liu et al. | Medical Virology | 2018 |
| 82 | CD56 | HBV | The universal deficit of p38 MAPK elicitation in CD56+ populations of NK cells from HBeAg(−) CHB patients is a potential cell-dependent function of this pathway during HBV infection. | Bakarozi et al. | Viral Hepatitis | 2019 |
| 83 | NLRP3 inflamasome | HBV | The CD14 macrophage population interacts with and downregulates NLRP3 during HBV infections to reduce protective responses against the virus. | Jia et al. | Molecular Immunology | 2020 |
| 84 | PBMC | X protein | The HBV intra-host diversity during mother-to-child transmission: the X protein could be responsible in viral survival amongst neonates post transmission. | Lie et al. | Archives of Virology | 2020 |
| 85 | LAG-3 | HBV | LAG-3 expression levels were significantly higher in CD8+ T-cell populations in CHB patients than healthy individuals. | Ye et al. | Medicine | 2017 |
| 86 | Asialoglycoprotein | HBsAg | HBV-infected mothers and their HbsAg-positive neonates had high expression of an asialoglycoprotein receptor isoform on dendritic cells. | Vyas et al. | Viral Hepatitis | 2018 |
| 87 | IL-21-vertical transmission | HBV | Mothers inflicted with HBV had low serum IL-21 levels and decreased TFh-cells and plasma B-cell concentrations, which were linked to vertical transmission to neonates. | Vyas et al. | Hepatology communications | 2018 |
| 88 | TfH | HBV | Mothers inflicted with HBV had low serum IL-21 levels and decreased TFh-cells and plasma B-cell concentrations, which were linked to vertical transmission to neonates. | Vyas et al. | Hepatology Communications | 2018 |
| 89 | Fas | HBV | The transcriptome and protein expression levels of apoptotic receptors FAS, CASP3, CASP8, and CASP9 in the HBV group were significantly higher than those in the healthy group. | Guo et al. | Experimental and Therapeutic Medicine | 2017 |
| 90 | CASP3 | HBV | The transcriptome and protein expression levels of apoptotic receptors FAS, CASP3, CASP8, and CASP9 in the HBV group were significantly higher than those in the healthy group. | Guo et al. | Experimental and Therapeutic Medicine | 2017 |
| 91 | CASP8 | HBV | The transcriptome and protein expression levels of apoptotic receptors FAS, CASP3, CASP8, and CASP9 in the HBV group were significantly higher than those in the healthy group. | Guo et al. | Experimental and Therapeutic Medicine | 2017 |
| 92 | CASP9 | HBV | The transcriptome and protein expression levels of apoptotic receptors FAS, CASP3, CASP8, and CASP9 in the HBV group were significantly higher than those in the healthy group. | Guo et al. | Experimental and Therapeutic Medicine | 2017 |
Hepatitis B virus (HBV) is a model virus that demonstrates hepatotropic characteristics. It can establish a chronic viral infection in humans through host immune anergy; however, therapeutics and vaccinations that focus on the liver have major limitations and in certain cases, reinfection can occur after liver transplant.65 This prompted us look for other factors involved in the pathogenesis of HBV that could be contributing to the pathogenesis and treatment resistance.
The extra-hepatic behavior of HBV is quite unusual. Therefore, understanding its dynamic interactions with host white blood cells could help describe the wide range of clinical manifestations. There is evidence to support the presence immune dysfunction during the infection and the deregulation of major cellular pathways during viral pathogenesis has also been studied quite extensively.66–68 Nonetheless, data of the HBV viral interactome in host white blood cells are limited and we still lack a fundamental understanding of host viral protein-protein interaction behaviors during HBV pathogenesis. Therefore, we extracted the available protein data pertaining to virus interactions with lymphocytes and categorized these results into three modules. These modules discussed the various aspects of HBV interactions with white blood cell proteins. We were able to draw a parallel between HBV and other viruses like hepatitis C (HCV) and HIV. HCV lymphotrophism is studied more extensively than HBV; whereas HIV is a lymphotropic virus that has a retroviral mode of propagation like HBV. HBV in vitro studies are difficult to conduct; therefore, molecular biology approaches aid in the validation of host viral interaction patterns. Additionally, comparative analysis of other viral pathogenic mechanisms can provide a direction for targeted research towards HBV lymphotrophism. A multi-dimensional analysis of the data retrieved from keyword-based text mining illustrated that a lymphotropic behavior of HBV should not be ruled out. Moreover, there are certain cellular pathways that may aid viral propagation in immune cells. Thus, the immune system may be contributing HBV expansion rather than its elimination.
DRYAD: Assessment of hepatitis B viral lymphotrophism using deep curation.
(DOI): doi:10.5061/dryad.qz612jmj1
This project contains the following underlying data:
• Data file 1. python_script: python code used for text mining from PUBMED
• Data file 2. hbv_keywords_1: List of keywords used for text mining from PUBMED.
• Data file 3. HBV_TEXT_MINING_MASTER_LIST: It is the excel list of all the publications extracted from PUBMED with respect to the keywords used for text mining.
• Data file 4. FINAL_HBV-WBC_CURATED_MOLECULES_WITH_CITATIONS: the filtered publication data from the master list pertaining specifically to the HBV and white blood cells.
• Data file 5. FINAL_HBV-WBC_CURATED_MOLECULES_WITH_CITATIONS_MOLECULE_LIST: the further filtered list of all the host white blood cells or molecules where the evidence of HBV replication has been found.
• Data file 6. FINAL_HBV-WBC_CURATED_MOLECULES_WITH_CITATIONS_MOLECULE_LIST: The list of all the genes of white blood cells found to be involved during the infection.
• Data file 7. FINAL_HBV-WBC_CURATED_MOLECULES_WITH_CITATIONS_PROTEIN_LIST: The list of all the host white blood cells proteins found to be involved during HBV infection.
• Data file 8. FINAL_HBV-WBC_CURATED_MOLECULES_WITH_CITATIONS_PBMC: The evidence of general trend of PBMCs behavior during the HBV infection.
• Data file 9. F1000_research-Further_reading: List of publication to supplement the information of the text mining data.
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
I would like to thank the team of Rawal Labs at Amity university Noida for supporting this work through their valuable inputs.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)