Keywords
Extraction technique, Phenolic compounds, Phytochemical, Syringodium isoetifolium, Ultrasound
This article is included in the Agriculture, Food and Nutrition gateway.
Extraction technique, Phenolic compounds, Phytochemical, Syringodium isoetifolium, Ultrasound
Indonesia's vast coastline is a habitat for seagrass, which grows to form seagrass meadows. Syringodium isoetifolium, Thalassia hemprichii, Cymodocea rotundata, Halophila ovalis, Enhalus acoroides, Cymodocea serrulata, Halodule pinifolia, and Halodule uninervis are species that are often found in seagrass beds in Indonesian waters (Kiswara et al., 2009; van Katwijk et al., 2011). The species S. isoetifolium, Cymodocea rotundata, and Cymodocea serrulata have a growth period that tends to be faster (Kiswara et al., 2009). Recently, Li et al. (2021) succeeded in seeding E. acoroides for the restoration of seagrass beds in the tropics. Therefore, seagrass is a marine plant that has the potential to be produced on a large scale. However, the high production potential of seagrass has not been followed by its postharvest utilization to produce high economic value products. So far, seagrass beds have been allowed to grow wild and play an important role not only in coral reef ecosystem and maintain the sustainability of the marine biota, but also in maintaining other marine ecosystem’s balance (van Katwijk et al., 2011).
Only the leaves of seagrass that has optimal growth can be harvested so that it does not cause the plant’s death. In addition, there is no need to replant after harvesting seagrass leaves. Seagrass leaves have a dark green color and a strong aroma, and are rich phytochemical compounds such as phenolics. Phenolic compounds are molecules with high diversity formed by the attachment of one or more –OH groups to the phenyl ring. Phenolics are divided into several classes including flavonoids and other benzopyran derivatives, phenolic acids and ester derivatives, quinones, lignins, lignans, tannins, stilbenes, curcuminoids, chalcones, and essential oils (Chiorcea-Paquim et al., 2020).
As seagrass has a deep green color and a quite strong aroma, it is predicted that it is rich in phytochemical compounds such as phenolics. A previous study by Rengasamy et al., 2019 reported that S. isoetifolium has a potential as a natural antioxidant and clinical enzyme inhibitor that can be applied in pharmaceutical cosmeceutical industries. Moreover, S. isoetifolium could also be a good candidate as a food complement, feed, and biomedical field due to its rich biochemical profile (Bharatharathna & Santhanam, 2019).
The extraction technique plays an important role in extracting nutrient content and phytochemical constituents. Efficient extraction techniques that are fast, inexpensive, economical in materials and solvents, high-yield, environmentally friendly, and do not damage the structure and functional properties of the target compounds, were chosen. Conventional extraction is usually carried out by maceration, which involves immersing the material with a solvent at a certain temperature for a certain time with agitation assistance (Abd Aziz et al., 2021); meanwhile, modern extraction techniques are carried out using microwave-assisted extraction (MAE) and ultrasonics. The principle of MAE is to use energy from microwaves to heat a mixture of sample material and extraction solvent, which is induced by microwave irradiation (Lefebvre et al., 2021). Microwave power is strongly absorbed by the interior of plant cells due to their water content. The pressure on the cell wall is generated by swelling of the plant cell when water evaporates, which pushes the cell wall from within to stretch and eventually rupture. This event can release metabolites from the broken cells into the surrounding solvent (Bachtler & Bart, 2021). On the other hand, the principle of UAE is to use energy from ultrasonic waves. At a certain frequency and amplitude, it creates cavitation bubbles, which when they reach an unstable point, then release high temperatures and pressures by the blasting process. This phenomenon can damage cell walls and promote the release of metabolites from the plant (Lefebvre et al., 2021). These different extraction techniques affect the quantity and bioactivity of the target compounds that are crucial to study.
To our knowledge, a recent study by Kalaivani et al. (2021) has reported the biological activities such as antibacterial, antifungal, antimicrobial, antifouling, and anticancer properties of S. isoetifolium. This research was conducted to develop the potential use of S. isoetifolium as a food bioactive contituents. To get the best phytochemical and nutrient compounds from S. isoetifolium, four different techniques, namely maceration, MAE, and UAE-bath (UAE with bath) and UAE-probe system were applied. Meanwhile, in this study, the phenol extract was analyzed and phytochemical compounds were screened by liquid chromatography–high resolution mass spectrometry (LC-HRMS). While the potential development is proven by in silico study and cell toxicity.
All experiments were carried out at Universitas Brawijaya, Indonesia. Fresh S. isoetifolium was harvested from Kondang Merak Beach, East Java, Indonesia (-8.3964160388196N, 112.51923979683157E) and then directly washed with fresh water to remove attached dirt and salt particles. Afterward, the seagrass was dried using an oven at 40°C to a constant weight (13.87 ± 0.20% moisture) and then with a blender. The dried powder (297.00 μm) was used for extraction. Chemicals used for extraction and analysis comprised 2,2,1-diphenyl-1-picrylhydrazyl (DPPH), Folin–Ciocalteu phenol reagent, sodium carbonate (Na2CO3), gallic acid, Aquadest, and absolute ethanol, from Meck, Singapore. All chemicals used were analytical grade except water and ethanol for LC-HRMS analysis, which were HPLC-grade.
Preparation
Water, ethanol 50%, and ethanol 100% were applied for extraction with different techniques. So far, they have been considered green solvents because they have been proven safe (non-toxic) for either phytochemical extraction or food usage (Ahmad et al., 2021; Molino et al., 2018). In addition, green solvents have a positive environmental impact due to their lower eco-toxicity and are biodegradable due to a strong solvency (Ahmad et al., 2021). Also, these solvents are highly efficient to completely extract the molecules in various polarities (Farooq et al., 2021; Harscoat-Schiavo et al., 2021; Lao et al., 2017).
Extraction process
Seagrass powder in each extraction solvent (viz. water, ethanol 50%, and ethanol 100%;1:20 w/v) was extracted with different techniques, i.e. MAE (Anton Paar - Multiwave Pro), UAE-Probe (Branson 250/450), UAE-Bath (Soltec - Sonica 2400 EP Ultrasonic cleaner) and conventional maceration (Waterbath shaker – Memmert WNB 45). MAE followed the Albuquerque et al. (2017) method with a slight modification in extraction time. MAE settings were: 50°C, stirrer at high speed, a 5 min duration, and factory set (Maximum microwave power 700 W; Maximum pressure 18.0 bar; Maximum pressure increase rate 0.5 bar/s). UAE-Bath followed the optimum method by Albuquerque et al., 2017) with the following equipment settings: off-temperature control, 60 min duration, and factory set (40 kHz frequency, 260 W peak ultrasonic power). UAE-Probe followed (Albuquerque et al., 2017) method by modifying the maximum time. UAE-Probe settings were: 70% amplitude, off-temperature control, 5 min duration, continuous duty cycle, and factory set (19-20 kHz horn frequency, 400 W kinetic energy). Conventional based on the optimum method by (Albuquerque et al., 2017), extraction was processed in a water bath shaker, agitated at medium speed, at 30°C for 1440 min. Each treatment was done in triplicate.
Dry extract generated
The extracted mixture was filtered with filter paper and then centrifugated at 3000 rpm, 25°C, for 5 minutes to separate the solute and the material residue. The extract solution was dried with a vacuum oven at 40°C to achieve constant weight. The water, 50% ethanol, and 100% ethanol dry extracts were yielded for each extraction technique. The dry extracts were then stored at -20°C for the next analysis. The yield of dry extracts was calculated using Equation ((1) (Aboulghazi et al., 2022).
Analysis of total phenolic content followed a Folin-Ciocalteau method (Lim et al., 2019). Briefly, 30 μl of Folin-Ciocalteu reagent (1.0 N) and 60 μL of diluted samples were mixed in a 96-well microplate, then incubated for 5 min. Afterward, 150 μL of 20% sodium carbonate solution was added and incubated in a dark place at room temperature for 40 min. The absorbance of the supernatant was measured at 730 nm using Microplate Reader after centrifugation for 8 min (1600×g). The total phenolic content of the samples was calculated with a standard curve using gallic acid (mg GAE/g w.b.).
Identification of phenolic compounds contained in 50% ethanol extract of S. isoetifolium using different extraction techniques was carried out using a non-targeted screening method (Susilo et al., 2020) by dissolving 100 μL of seagrass extract in 1400 μLof water and ethanol depending on the solvent used for extraction. Samples were filtered with a 0.22 m minisart RC and then injected into the LC-HRMS apparatus. The 10 μL samples were processed automatically by using a hypersil gold aQ 50 mm × 1 mm × 1.9 μm (length x diameter x particle size) column with positive polarity conditions, flow rate 40 L/min, oven column temperature 30°C with elution gradient as follows: 0–2 min 5% B, 15–22 min 60% - 95% B, stabilized at 95% B for 3 min then dropped to 5% B for 5 min. The chromatogram data generated from the injection process was analyzed using Compound Discoverer 3.2 software based on the Mzcloud online library.
Ligand and protein preparation
The ligands used in this study consisted of choline compounds and inhibitors of each protein. All ligands were obtained from the PubChem web server in Sybil Data Files (SDF) format. The ligands were converted into a 3D structure in Protein Data Bank (PDB) format with BIOVIA Discovery Studio 2019. Choline is known to have anti-inflammatory activity, and in this study, it was hoped that it could bind to TNF-α, IL-1β, and IL6, which function as pro-inflammatory agents. The 3D structures of TNF-α (2AZ5), IL-1β (1ITB), and IL6 (1P9M) were obtained from the RCSB PDB web server. The receptors were then prepared to remove water molecules and ligands with BIOVIA Discovery Studio 2019.
Molecular docking analysis and ligand-receptor interactions
Ligand-receptor interactions were analyzed by molecular docking using AutoDock Vina integrated in PyRx version 0.9.5. The molecular docking process was carried out using a specific docking method based on the active site of each receptor (Boulanger et al., 2003; Halim et al., 2015; He et al., 2005; Zia et al., 2020). Docking results, bond positions, and amino acid residues formed between the ligand-receptors were analyzed using the 2019 BIOVIA Discovery Studio.
Molecular dynamic simulation
Molecular dynamic (MD) was used as a simulation method to analyze the physical movements of atoms and molecules. The Yet Another Scientific Artificial Reality Application (YASARA) has been used for MD simulations. This simulation was carried out to compare the interaction of ligands and inhibitors in binding to receptors. The parameters in the simulation correspond to the physiological conditions of the cell at 37°C, 1 atm, pH 7.4, and 0.9% salt content for 50 ns with autosave every 25 ps. The simulation was run by the md_run macro program, and the results were displayed by the md_analyze and md_analyzebindenergy programs (Bagheri & Fatemi, 2018; Deeba et al., 2017; Krieger & Vriend, 2015).
In this study, the MTT assay based on Miasih et al. (2022) was applied with slight modification. The extract of S. isoetifolium was treated in TIG-1 cells. Finally, the absorbance was measured using a microplate reader at a wavelength of 595 nm. The results were used to construct a graph of cell viability percentage against extract concentrations.
Using 100% ethanol, 50% ethanol, and water as solvents aims to extract phytochemical compounds with high polarity to low polarity. The extraction results are presented in Table 1, the water and 50% ethanol solvent produced the highest yield while 100% ethanol solvent produced a significant yield for each extraction technique. It was expected, due to the phytochemical compounds (such as dihydrocaffeic acid, quercetin, caffeic acid, levalbuterol, phloretin, and zearalenone; see in Table 3) of S. isoetifolium having high polarity. In other plants (Blepharis linariifolia Pers., Guiera senegalensis JF Gmel. and Limnophila aromatica), 50% ethanol solvent also showed a higher yield than 100% ethanol, 75% ethanol, water, acetone, and dichloromethane (Dirar et al., 2019; Do et al., 2014). Many studies have stated the advantages of non-conventional extraction methods compared to conventional ones, namely high yields (Ameer et al., 2017). However, the yield of S. isoetifolium extraction with various non-conventional and conventional techniques showed no significant difference. The extraction yields depends on the extraction time (Đurović et al., 2018).
Phenolics are molecules with high diversity defined by the attachment of one or more –OH groups to the phenyl ring. Phenolics are divided into several classes including flavonoids and other benzopyran derivatives, phenolic acids and ester derivatives, quinones, lignins, lignans, tannins, stilbenes, curcuminoids, chalcones, and essential oils (Chiorcea-Paquim et al., 2020). For each extraction technique, extraction of S. isoetifolium with 50% ethanol as a solvent produced the highest total phenol, meanwhile, with 100% ethanol solvent, phenol could not be obtained (Table 1). The polarity of the phenolic compounds is medium so extraction with 50% ethanol in many plants (Moringa oleifera L. leaves, rambutan peel, Cyperus rotundus L., and pomegranate) yielded the highest total phenol compared to extraction with 100% ethanol, 80% ethanol, 70% ethanol , water, acetone, and dichloromethane (Dirar et al., 2019; Harscoat-Schiavo et al., 2021; Phuong et al., 2020; Wu et al., 2020). The total phenol content in S. isoetifolium extract was influenced by the extraction technique used. UAE-probe yielded a significantly higher total phenol (17.37 ± 2.16 mg GAE/g) compared to UAE-bath, MAE, and conventional (Table 1). Đurović et al. (2018) reported that the total phenol content of yellow soybean seeds extracts, which was extracted using ultrasound-probe, was not different from that of ultrasound-bath and MAE. This is due to the different samples extracted, the structure of yellow soybean seeds being harder than S. isoetifolium leaves, as well as the different processing conditions of the ultrasound-probe (20 kHz, 30% of maximum amplitude, 10 min duration, the temperature was held constant at 25 ± 1°C) and UAE-probe (50/60 kHz, 70% amplitude, 5 min, at room temperature and the temperature was increased but not held).
Phenolic compounds are non-polymeric phytochemical compounds that present bioactivity as antioxidants. Their main structure is an aromatic ring or phenol unit containing at least one hydroxyl group (Akyol et al., 2016; Lao et al., 2017; Yang et al., 2018). The complex phenolic structure causes a wide variety of molecular weights, and there are about 10,000 different phenolic derivatives (Chiorcea-Paquim et al., 2020). Caffeic acid, quercetin, coumarin, vanillic acid, benzoic acid, cinnamic acids, phloretin, vanillin, and benzopyran derivatives are examples of phenolic compounds (Akyol et al., 2016; Chiorcea-Paquim et al., 2020; Xiong et al., 2019). In this study, the extraction of S. isoetifolium with 50% ethanol produced the highest total phenol; therefore the use 50% ethanol for each extraction technique identified its phenolic compounds content. As can be seen in Table 2, UAE-probe yielded the most phenolic compounds (10 types), including 3,5-di-tert-Butyl-4-hydroxybenzoic acid, 7-hydroxycoumarine, 4-methoxycinnamic acid, zearalenone, caffeic acid, levalbuterol, phloretin, dihydrocaffeic acid, quercetin-3β-D-glucoside, and quercetin. Meanwhile UAE-bath, MAE, and conventional extraction yielded fewer phenolic compounds: 5, 5, and 6 types, respectively. Specifically, zearalenone, caffeic acid, phloretin, dihydrocaffeic acid, and quercetin were detected only by UAE-probe. The advantages of the UAE for the extraction of phenolic compounds have also been proven in other plants. The yield of ellagic acid and quercetin extraction from rambutan peel with UAE was significantly higher than conventional extraction (Phuong et al., 2020). Extraction yield of quercetin, gallic acid, vicenin-2, p-Hydroxybenzoic acid, orientin, rutin, hyperoside, kaempferol-3-O-rutinoside, isorhamnetin 3-O-glucoside, rosmarinic acid, apigenin, and kaempferol on Moringa oleifera L. Leaves with UAE was higher than with conventional extaction (maceration and stirring-assisted extraction) (Wu et al., 2020). Interestingly, choline was abundantly present in all extraction techniques, especially with UAE-probe. Choline is an essential nutrient that plays an important role for neurotransmitter synthesis (acetylcholine), cell-membrane signaling (phospholipids), lipid transport (lipoproteins), and methyl-group metabolism (homocysteine reduction). Moreover, choline is also required to make the phospholipids phosphatidylcholine, lysophosphatidylcholine, choline plasmalogen, and sphingomyelin – essential components for all membranes (Zeisel & da Costa, 2009). Betaine also presents in all extraction techniques, except MAE. According to findings from a previous study by Cho et al. (2006) and Detopoulou et al. (2008), a combination of dietary intake rich in choline and betaine are associated with lower concentrations of all of the inflammatory markers, including C-reactive protein (CRP), homocysteine, interleukin-6, and tumor necrosis factor.
Ligand-receptor interactions
Molecular docking is used to determine the interaction between the receptor and the ligand, indicated by the binding affinity value and the amino acid residues involved (Figure 1). Since the presence of choline is abundant, this study used a ligand consisting of choline compounds and inhibitors from each protein. The inhibitor of each protein is a compound used as a control, to compare the strength of the interaction between choline-receptors and inhibitor-receptors. Molecular docking results show that the interaction of choline compounds with receptors (TNF-α, IL-1β, and IL6) had a higher binding affinity value than the inhibitor-receptor interaction, namely -3.4, -3.0, and -2.8 kcal/mol. The binding affinity between the inhibitor and TNF-α was the highest compared to the other inhibitor-receptors, with a value of -9.1 kcal/mol. The higher the binding affinity value, the weaker the bond between the ligand-receptor (Table 3).
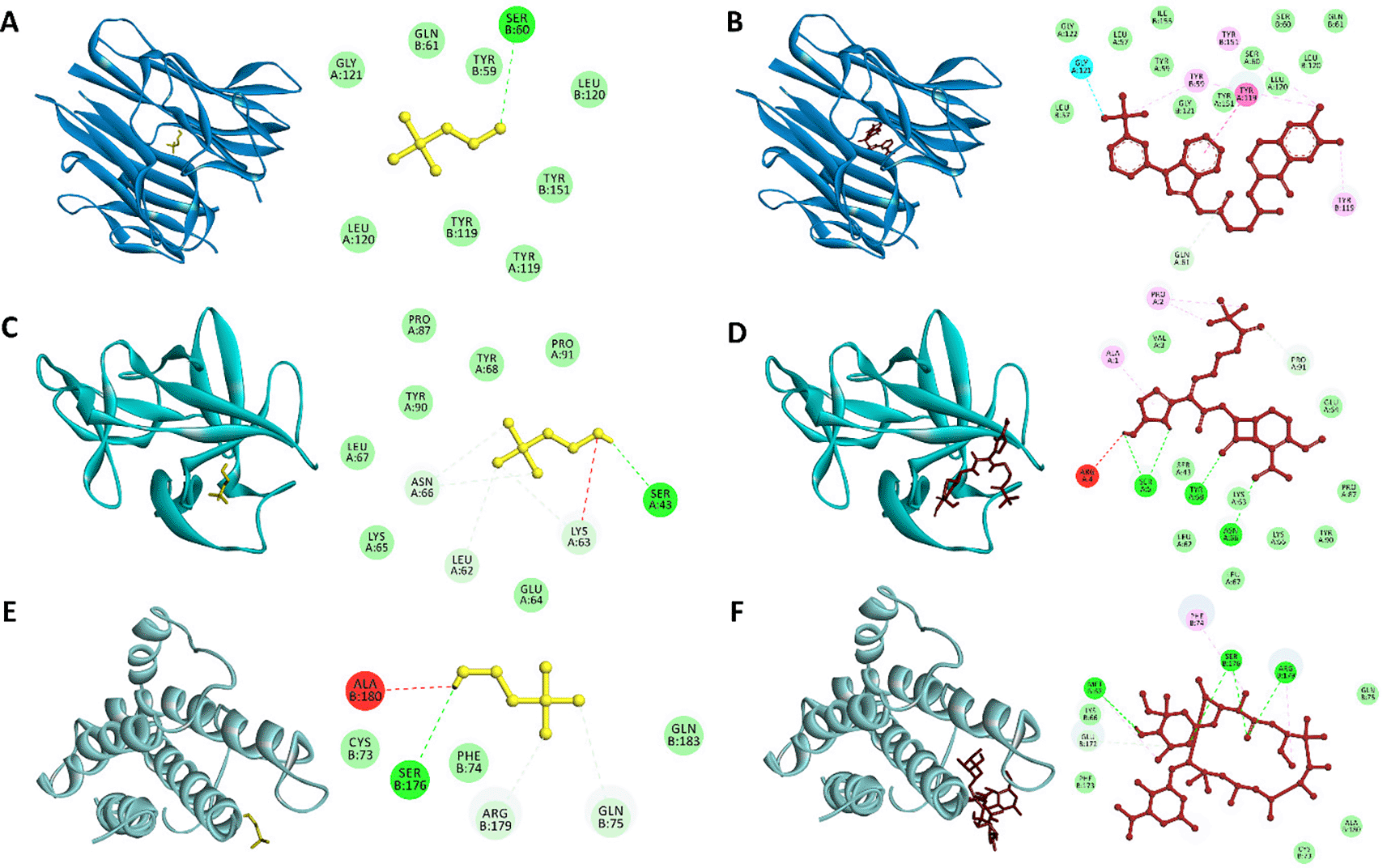
(A) Choline-TNF-α, (B) Inhibitor-TNF-α, (C) Choline-IL-1β, (D) Inhibitor-IL-1β, (E) Choline-IL6, and (F) Inhibitor-IL6. Choline and inhibitors are presented as yellow and red sticks, respectively. TNF-α, IL-1β, and IL6 are presented as ribbon structures with the different blue color.
Molecular docking results were then visualized to determine the binding site of the ligand and receptor. Visualization with Discovery Studio showed that the choline compound had the same binding site as the inhibitor on each protein. The presence of the same binding site indicates that the choline compound can have the same function as the control, even though it has a relatively high binding affinity value. The same binding site between choline compounds and inhibitors in each protein was indicated by several amino acid residues that interact through hydrogen and hydrophobic interactions. Tyr59, Ser60, Gln61, Tyr119, Leu120, Gly121, and Tyr151 are seven amino acid residues involved in the interaction of choline-TNF-α and inhibitor-TNF-α. However, there was a slight difference: Ser60 in the choline-TNF-α reaction interacts through hydrogen bonds, whereas in the inhibitor-TNF-α reaction, it interacts through hydrophobic bonds. Tyr119 is an amino acid residue that plays an essential role in ligand-receptor interactions (Zia et al., 2020).
Leu62, Lys63, Glu64, Lys65, Asn66, Leu67, Tyr68, Pro87, Tyr90, and Pro91 are 10 amino acid residues involved in the interaction between choline-IL-1β and inhibitor-IL-1β. Asn66 and Tyr68 are two amino acid residues that interact through hydrogen bonds in the inhibitor-IL-1β interaction while interacting through hydrophobic bonds in the choline-IL-1β interaction. Ser176 is an amino acid residue that interacts via hydrogen bonds in the reaction of choline-IL6 and inhibitor-IL6. Four other amino acid residues involved in the reaction of the two ligands with IL6 interact via hydrophobic bonds, namely Cys73, Phe74, Gln75, and Ala180. Amino acid residues involved in the ligand-receptor interaction complex through hydrogen bonds and hydrophobic bonds can increase the affinity of the ligand bonds in proteins (Chen et al., 2016). It is known that the formed hydrophobic interactions in ligand-receptor complex play an important role in stabilizing the ligand-protein bond and helping to increase the affinity of the ligand bond with protein (Varma et al., 2010).
Molecular dynamic simulation results
The stability of the ligand complex with each pro-inflammatory protein is known based on molecular dynamic (MD) simulations. As seen in Figure 2, the parameters used in this simulation were the RMSD of the ligand-receptor complex, RMSF, and the number of hydrogen bonds of the ligand-receptor complex. MD analysis showed that the choline-TNF-α complex and the inhibitor-TNF-α had the same stability up to 20 ns simulation, indicated by an RMSD value of 3 Å. The stability of the choline-TNF-α complex fluctuated with RMSD values, reaching 3.5 Å at several simulation times. Unlike the inhibitor-TNF-α complex, which experienced relatively high fluctuations during the simulation time of around 29 ns with an RMSD value of 4 Å, the simulation stabilized again after 30 ns with an RMSD value of less than 3 Å. The simulation of the choline-IL-1β complex was very unstable, as indicated by very high fluctuations and an average RMSD value of more than 4 Å. These results are inversely proportional to the stable inhibitor-IL-1β complex from start to end of the simulation, indicated by the average RMSD value <3 Å. The choline-IL6 and inhibitor-IL6 complex was unstable compared to other interactions, indicated by an RMSD value of >3 Å. However, when paying attention to the choline-IL6 complex, it had a higher level of stability than the IL6-inhibitor, indicated by a lower RMSD value. Previous studies have shown that the ligand-receptor complex, which is stable during simulation, has an RMSD value of 3 Å (Martínez, 2015; Wargasetia et al., 2021).
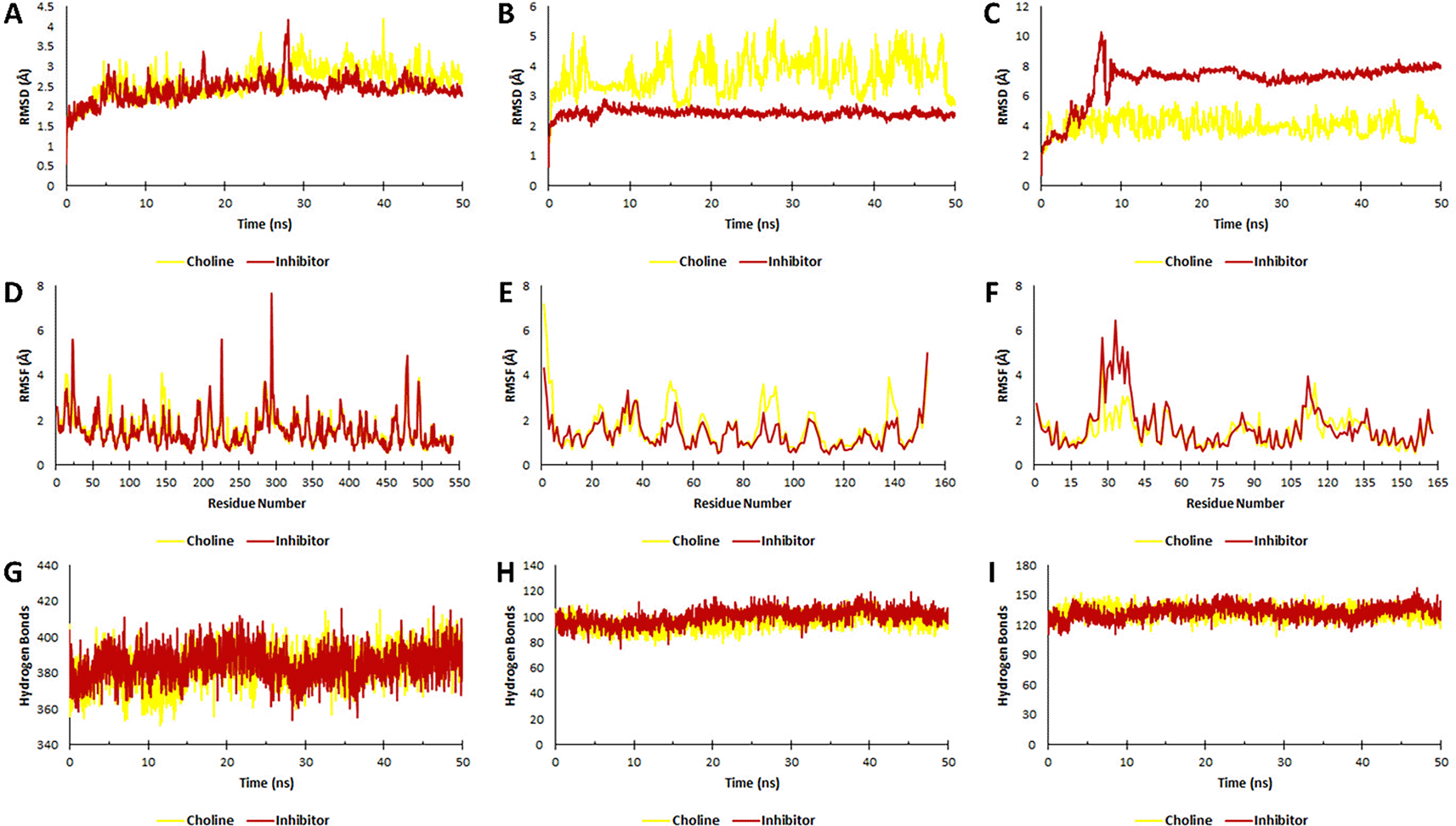
During simulation, the stability of the ligand-protein interaction complex is indicated by the RMSD value. RMSF value is based on fluctuations in the amino acid residues and the hydrogen bond numbers. A, D, G. Ligands-TNF-α complex. B, E, H. Ligands-IL-1β complex. C, F, I. Ligands-IL6 complex.
The fluctuations of the amino acid residues formed can also determine the stability of the ligand-receptor complex. The choline-TNF-α and inhibitor-TNF-α complexes are the most stable compared to other ligand-receptor complexes. This is indicated by fluctuations in amino acid residues that are not too high compared to the others. It is known that the more amino acid residues that experience fluctuation, the more unstable the ligand-receptor complex (Azminah et al., 2019). The number of hydrogen bonds in the choline-TNF-α and inhibitor-TNF-α complexes were the highest compared to the other complexes. A large number of hydrogen bonds makes the two ligand-receptor complexes the most stable compared to other ligand-receptor complexes.
A toxicity test was conducted to evaluate the effect of the extract on the viability of TIG-1 cells. TIG-1 cell viability was determined by the MTT-assay method, using mitochondrial dehydrogenase activity in living cells against 3[4,5-dimethylthiazole-2-yl]-2,5-diphenyltertrazolium bromide (MTT) into insoluble purple formazan crystals. The MTT test requires the addition of a solvent (DMSO) to dissolve the formed insoluble formazan product.
The data obtained were the absorbance values of purple formazan crystals measured at a wavelength of 595 nm, which were then converted to determine the percentage of living cells. The number of viable cells was measured by colorimetry and the quantity of formazan product was directly proportional to the number of live cells in the culture (McCauley et al., 2013).
The results of the toxicity test (Figure 3) showed a decrease in the viability of TIG-1 cells in line with the increase in the concentration of the extract, but not significant. Exposing the lowest extract concentration of 12.5 μg/mL showed no decrease in TIG-1 cell viability compared to control cells, which was 96.1%. Meanwhile, exposing the highest extract concentration of 800 μg/mL showed a decrease in cell viability of 78.4%. TIG-1 cell viability in the range of extract concentrations of 25-400 μg/mL did not show a significant difference in the range of 84.5-92.5%. The results of the extract toxicity test on TIG-1 cells showed that the extract did not have a toxic effect on TIG-1 cells.
Referring to international guidelines such as those of the International Organization for Standardization (ISO 10993-5:2009) it has been shown that if the viability is reduced to <70% of the control, then the test substance has the potential for cytotoxicity. Based on these guidelines, giving the extract to TIG-1 cells with the highest dose has a viability value above 70% which indicates that the extract has no cytotoxic potential on normal cells.
An in vitro cytotoxicity test was used to evaluate the toxicity of extracts following international guidelines such as those by the International Organization for Standardization (ISO 10993-5:2009). This test can be used to determine the structure–activity relationship of the extract used, as well as determine the initial concentration for further research. Inhibition of cell death, cell growth, or cell proliferation are important parameters used to evaluate the cytotoxicity of an extract (Aydin et al., 2016).
Bioactive constituents of Syringodium isoetifolium can be extracted efficiently using the UAE-probe method with 50% ethanol; in this study, it resulted in both the highest yields and total phenol, as well as the richest bioactive constituents. Moreover, choline was the most dominant compound in extracts yielded by different extraction techniques. Since choline was an abundant compound, the in silico assay was done using this compound, and resulted in choline having the ability to be anti-inflammatory. Furthermore, based on a cytotoxicity test, the S. isoetifolium extract by UAE-Probe with 50% ethanol did not have a toxic effect on TIG-1 cells. Hence, Syringodium isoetifolium extract had a high potential as a bioactive food ingredient.
Figshare: Experimental Data: Potency Development of Seagrass (Syringodium isoetifolium) as Food Bioactive Constituents by Different Extraction Techniques, https://doi.org/10.6084/m9.figshare.21780674.v7 (Susilo et al., 2022).
This project contains the following underlying data:
Figshare: Experimental Data: Potency Development of Seagrass (Syringodium isoetifolium) as Food Bioactive Constituents by Different Extraction Techniques, https://doi.org/10.6084/m9.figshare.21780674.v7 (Susilo et al., 2022).
This project contains the following extended data:
- Instruments.xlsx
- Minitab Project - ANOVA OF YIELD DATA.MPJ.BAK
- Minitab Project - ANOVA of Yield Data.MPJ
- Minitab Project - ANOVA OF TOTAL PHENOLIC DATA.MPJ.BAK
- Minitab Project - ANOVA of Total Phenolic Data.MPJ
- GraphPad Prism - Graphic of TIG-1 cell viability profile.pzfx
- Table. Molecular docking and amino acid residues on ligand-receptor interactions.docx
- Figure. Molecular dynamic simulation.png
- Figure. Ligand-receptor interaction and amino acid residues involved.png
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
The authors are grateful to research center and community services (LPPM); central laboratory of life sciences (LSIH), Faculty of Agricultural Technology, Universitas Brawijaya, Malang-Indonesia for the funding and the instrumentation support.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Extraction of bioactives by emerging green methods, especially by supercritical fluids.
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Emerging technologies Extraction Processes
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 31 Aug 23 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)