Keywords
Aldose reductase, Microvascular complications, Chimera, SWISSADME, Luteolin, Quercetin, Sulindac
This article is included in the Cheminformatics gateway.
Aldose reductase, Microvascular complications, Chimera, SWISSADME, Luteolin, Quercetin, Sulindac
Diabetes mellitus (DM) is the number nine leading cause of death globally and a major reason for the increase in the number of male deaths since the year 2000 (World Health Organization, 2020). Currently, approximately half a billion people worldwide have been diagnosed with DM and cases are projected to rise above 600 million by 2045 (Saeedi et al., 2019). Furthermore, diabetes predisposes patients to developing other morbid conditions such as hypertension. Despite these facts, diabetes especially type 2 DM, is manageable and preventable. By definition, DM is a chronic metabolic disease that once diagnosed with, has a poor recovery prognosis (Sun et al., 2022). Its chronic form is associated with the development of medical emergencies (diabetic ketoacidosis (DKA), Hyperosmolar Hyperglycemic State (HHS)), progressive microvascular (retinopathy, neuropathy, nephropathy) and macrovascular (ischemic heart disease, peripheral vascular disease, stroke) complications that reduce the quality of life of patients (Chawla et al., 2016). Additionally, DM has been reported as being immunosuppressive as seen by increasing incidences of infections in patients suffering from it (Polk et al., 2021; Dryden et al., 2015, Hobizal & Wukich, 2012, Devrajani et al., 2010). Diabetes also increases the risk of developing other non-communicable conditions such as chronic kidney disease and dyslipidemias.
In diabetic complications, the pathological hallmark commonly surrounds the vasculature system leading to the development of microvascular and macrovascular complications (Eid et al., 2019). These complications are progressive in nature and with poor prognosis. Studies such as the United Kingdom Prospective Diabetes Study, reports that strict control of blood glucose levels limits microvascular disease but use of glucose-lowering agents relatively improves macrovascular outcomes (King et al., 1999). Background literatures portrays a mixed picture of development of diabetic complications; some scholars suggest microvascular and macrovascular complications occur simultaneously, while others posit that macrovascular complications occur independent of microvascular complications (Eid et al., 2019, Chawla et al., 2016). At the core of the cardiovascular system, microvessels (arterioles, capillaries, venules) form the basic functional unit. Blood moves between microvessels and macrovessels to supply cells with oxygen and nutrient and remove waste products. However, the cellular components and architecture of microvessels differ from that of macrovessels. Macrovessels primarily function as transport medium, while microvessels regulate blood pressure, control vascular permeability and optimize blood flow to the needs of the cells.
In patients with diabetes, excessive glucose due to hyperglycemia causes thickening of the capillary basement membrane and increased protein synthesis within the extracellular matrix. These changes together with advanced glycation end products (AGEs), and inflammation induces microangiopathy, which facilitates the development of microvascular complications, i.e., retinopathy, neuropathy and nephropathy (Giacco & Brownlee, 2010). Moreover, since these microvessels control blood pressure, such abnormalities lead to the development of hypertension. Thus it can be understood that macrovascular complications are independent risk factors for microvascular complications. However, there is a strong correlation between the two with other studies suggesting microvascular changes to be risk factors for macrovascular complications (Hurst et al., 2015). Common pathways leading to these complications are the formation of AGEs, induction of oxidative stress, low grade inflammation, neovascularization of vasa vasorum and the sorbitol pathway.
This study focuses on the sorbitol pathway/polyol pathway, which has been implicated in the development of microvascular complications (Yan, 2018). The development of microvascular complications follows a complex pathophysiology with inputs from several cellular biochemical pathways; one of them being the polyol pathway. This pathway does not solely and in absolution lead to the development of microvascular complications but has a major contribution and is critical for the pathogenesis of such complications as it has been observed in several studies (Yan, 2018; Mathebula, 2015; Lorenzi, 2007). The sorbitol pathway is a two-reaction step used to convert glucose to fructose as depicted in Figure 1.
Aldose reductase is a non-specific enzyme catalyzing the conversion of any sugar into its alcohol form. In healthy individuals, glucose affinity for aldose reductase is quite low but in hyperglycemic conditions, the increased glucose molecules in circulation increases enzyme affinity (Jannapureddy et al., 2021). The enzyme is found highly localized in specific cells such as epithelia of the lens, papilla and cortical cells in kidney, Schwann cells in peripheral nerves and islets of Langerhans in the pancreas. Ocular, neuronal, renal and pancreatic B-cells absorb glucose using insulin-independent glucose transporters (Jannapureddy et al., 2021). As such, the aforementioned conditions provide an ideal environment for microvascular complications to ensue. The intermediate product NADP+ acts as a negative feedback inhibitor of the enzyme preventing increased synthesis of sorbitol. However, the formed sorbitol is converted to fructose by sorbitol dehydrogenase—another non-specific enzyme leading to the final products of NADPH and fructose (Garg & Gupta, 2022). This negates the negative feedback of NADP+, thus ultimately, in equilibrium conditions, the forward reactions of the two reactions are favored. Within the specific cells, not all sorbitol is converted to fructose as the latter has not much use for cells in terms of energy production. Therefore, both fructose and sorbitol exist within the intracellular component of the cells yet they are osmotically active (Garg & Gupta, 2022).
Retinopathy; within the epithelial cells of the lens, sorbitol and fructose leads to swelling of the lens due to absorption of water attributed to osmotic effects of sorbitol and fructose. The swelling causes formation of hydropic fibers that with time liquefy due to continued swelling, resulting in intrafibrillar cleft formation. These intrafibrillar clefts lead to opacities and cataracts (Moemen et al., 2020). Neuropathy; diabetic neuropathy is pathologically characterized by segmental demyelination. Schwann cells are responsible for the synthesis of myelin compounds and their incorporation around nerves. By doing so, they increase nerve impulse conduction. The pathogenic pathway of diabetic neuropathy in relation to sorbitol levels is not clearly understood. It is postulated that the excess sorbitol and fructose causes swelling of the Schwann cells, which diminishes their functional capability and in the long run causes their death. This explains the noted improvement in peripheral function if hyperglycemia is managed in acute diabetic conditions and also the chronic permanent irreversible change associated with it (Ab Hamid et al., 2021). Nephropathy; although chronic diabetes is associated with the development of nephropathy, the exact mechanism is yet to be elucidated. However, accumulation of sorbitol and fructose has been implicated in its development especially on the papilla and cortical cells (Luis-Rodríguez, 2012).
Currently, over 600 plant species have been shown to have antidiabetic properties (Kayarohanam, 2015). Extract from these plants have phytochemical compounds that influence the glucose metabolic pathways and impart the pathways that lead to development of diabetic complications. The various phytochemicals work either independently or synergistically to exert their antidiabetic activity; thus we cannot say in absolution that they solely inhibit the polyol pathway. However, advanced technology has enabled isolation, characterization and visualization of the plant phytochemicals from the extracts. Besides, studies on the effect of such phytochemicals on the enzymes of the polyol pathway have reported specific phytochemicals with potent inhibitory effects on the enzymes. Aldose reductase has been the most targeted in studies as it is a rate-limiting step. Constituents such as quercetin, rosmarinic acid, nepetrin, mangiferin, luteolin, curcumin, ellagic acid, butein, and eugenol have potent aldose reductase inhibitory effects. Others such as ellagic acid and caffeic acid inhibit both aldose reductase and the sorbitol dehydrogenase enzymes, thus acting as sequential inhibitors. As such, this study aimed to virtually screen such phytochemicals for structurally similar compounds and assess their docking scores, pharmacokinetic profiles and toxicological profiles using computational methods as potential lead compounds.
This study was carried out in the School of Pharmacy, Kabarak University. An in silico study design was employed for this research. Based on the literature review, several plants have been shown to inhibit the development of microvascular complications. The search terms “aldose reductase”, “polyol pathway”, “aldose reductase inhibition”, “microvascular complications”, and “flavonoids” were used to search for literature from the following search engines: PubChem (RRID:SCR_004284) and Google Scholar (RRID:SCR_008878). The last search was performed in August 2021. Target prediction using the online tool, swisstargetprediction, was used to assess and validate whether phytochemicals from such plants do bind to aldose reductase. Two phytochemicals, luteolin and quercetin, were selected. The predicted probability for binding to aldose reductase for luteolin and quercetin was 1.00. Sulindac was used as a comparator for this study as it is an approved aldose reductase inhibitor.
Structure and canonical smiles of luteolin and quercetin were obtained from the PubChem website. The canonical smiles were used to screen online databases via the online tool SwissSimilarity for structurally similar compounds. ZINC database (RRID:SCR_006082) is an open access database with millions of chemical compounds that was selected for this study. The database is embedded within SwissSimilarity enabling the concomitant screening. Results of the screened compounds were downloaded as an excel file having canonical smiles of the various chemical compounds and their various similarity index. A total of 20 chemical analogs, for each phytochemical compound, with highest similarity index were isolated and used for further analysis. Canonical smiles of the 20 selected chemical compounds were drawn and “cleaned” using the online tool PubChem Sketcher version 2.4 and each downloaded as a MDL molfile. Each of the downloaded compound will again be optimized using the computer program Avogadro (RRID:SCR_015983) software and minimized using UCSF Chimera v1.16 (RRID:SCR_004097).
Structure of aldose reductase (PDB ID 3rx4) was obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) (RRID:SCR_012820). Non-standard amino acids present in the enzyme were removed using UCSF Chimera.
AutoDock Vina (RRID:SCR_011958) version 1.2.0 (Eberhardt et al., 2021; Trott & Olson, 2010) was used to dock the selected 20 analogs of each phytochemical to standard aldose reductase enzyme and their corresponding docking strength tabulated. The program is available as a computer application that can be embedded and run simultaneously with UCSF Chimera. Visualization of the complexes formed between ligand and target protein and their interaction was carried out using BIOVIA Discovery Studio v21.1.0.20298 (RRID:SCR_015651) (alternative AutoDock (RRID:SCR_012746), which is a free software that can be used)
SwissADME, an online web tool, was used to analyze the pharmacokinetic profile and synthesizability of the selected 20 analogs for each phytochemical. In addition to target prediction for side effects, the other toxicology analysis carried out involved running the analogs on Protox-II (RRID:SCR_018506) server, a web tool used to analyze toxicology of compounds.
Selected 20 analogs of each phytochemical compound analyzed were tabulated together with their similarity scores. Molecular docking scores, other target prediction, pharmacokinetic profiling, synthesizability and toxicological analysis results were also presented in form of tables, graphs and charts. Interpretation of the obtained data involved looking for compounds with: stronger affinities to increase efficacy, highly selective and less toxic to reduce side effects and better pharmacokinetic profile that can enhance dosing regimen and administration.
Table 1 (Otieno, 2022) shows the plant species that have been shown to have antidiabetic activity and the phytochemicals that are purported to have antidiabetic activity. In addition, Table 1 shows that both luteolin and quercetin, which can be obtained from chamomile and fenugreek respectively, were predicted to have 100% probability of binding to aldose reductase.
| Plant species | Phytochemical | Predicted probability |
|---|---|---|
| Matricaria recutita (Chamomile) | Luteolin | 1.00 |
| Trigonella foecum-graceum (Fenugreek) | Quercetin | 1.00 |
Figure 2 depicts the structures of luteolin, sulindac and quercetin. Notably both have a primary aromatic system associated with a carbonyl group (a thiocabonyl in the case of sulindac). In addition, luteolin and quercetin have a secondary aromatic ring that is substituted by polar groups. Sulindac also has a secondary aromatic ring system that is substituted with a fluorine and carboxylic group.
Table 2 describes the analytic results for luteolin and its ZINC analogues. Luteolin was predicted to have a slightly stronger binding affinity (-9.7) for aldose reductase enzyme than sulindac (-9.6). Eight out of the 20 analogues of luteolin had docking below both sulindac and luteolin with ZINC000004349582 having the strongest predicted binding strength (-10.5). Moreover, all ZINC analogues were ≥ 99.8% similar to luteolin. Luteolin did not violate the Lipinski rule of five and had a high GI absorption. Additionally, luteolin was predicted to neither cross the blood brain barrier nor be a substrate of p-glycoprotein (p-gp) (also known as MDR1, ABCB1 or CD243). Although luteolin was predicted to not inhibit CYP2C19 and CYP2C9, they inhibited CYP1A2, CYP2D6 and CYP3A4. On the contrary, the comparator inhibited all cytochrome enzymes analyzed except CYP1A2 and CYP2D6. All eight analogues of luteolin had similar pharmacokinetic profiles to their parent phytochemical compounds due to high structural similarity. On toxicological analysis, luteolin was predicted to be safer, falling under toxicology class 5 (LD50=3,919 mg/kg). Two out of the eight luteolin analogues also fell under toxicology class 3, while the rest were under class 5 with similar predicted LD50 values
GI, gastrointestinal; p-gp, p-glycoprotein; BBB, blood brain barrier.
Table 3 describes the analytic results for quercetin and its ZINC analogues. Quercetin was predicted to have a slightly stronger binding affinity (-9.7) for aldose reductase enzyme than sulindac (-9.6). Analysis of quercetin analogues showed that 14 out of the 20 selected ZINC compounds had better binding affinity than both parent phytochemical and the comparator. In particular, the compounds ZINC000000039111, ZINC000575623588, and ZINC000000057845 had the highest docking scores (-10.6). Furthermore, all 14 analogues were ≥ 99.7% similar to quercetin. Pharmacokinetic profile prediction quercetin and the comparator (sulindac) did not violate the Lipinski rule of five and had a high GI absorption. Quercetin was predicted to neither cross the blood brain barrier nor be a substrate of p-gp (also known as MDR1, ABCB1 or CD243). Quercetin did not inhibit CYP2C19 and CYP2C9 but was an inhibitor of CYP1A2, CYP2D6 and CYP3A4. To the contrary, the comparator inhibited all cytochrome enzymes analyzed except CYP1A2 and CYP2D6. All 14 analogues of quercetin had similar pharmacokinetic profiles as their parent phytochemical compounds due to high structural similarity. On toxicological analysis, quercetin was relatively unsafe, falling under toxicology class 3 (LD50=159 mg/kg). Five quercetin analogues had LD50 of 159 mg/kg, while another five had LD50=3,919 mg/kg. Two of the remaining four quercetin analogues had LD50=4,000 mg/kg, while the other two had a LD50 of 5,000 mg/kg. Quercetin analogues with highest predicted binding affinity: compound ZINC000000039111 and ZINC000575623588 had an LD50 of 159 mg/kg, while ZINC000000057845 had an LD50 of 4,000 mg/kg.
GI, gastrointestinal; p-gp, p-glycoprotein; BBB, blood brain barrier.
Figure 3 displays the pictorial representation of aldose reductase in a 3D model. Figure 3A shows the peptide chains (as different colors) that constitute the entire enzyme protein. Figure 3B displays the hydrophobic surface of the active binding site of aldose reductase. Figure 3C shows how the molecule sulindac fits into the binding pocket, while Figure 3D shows the bond interaction between sulindac and enzyme amino acids.
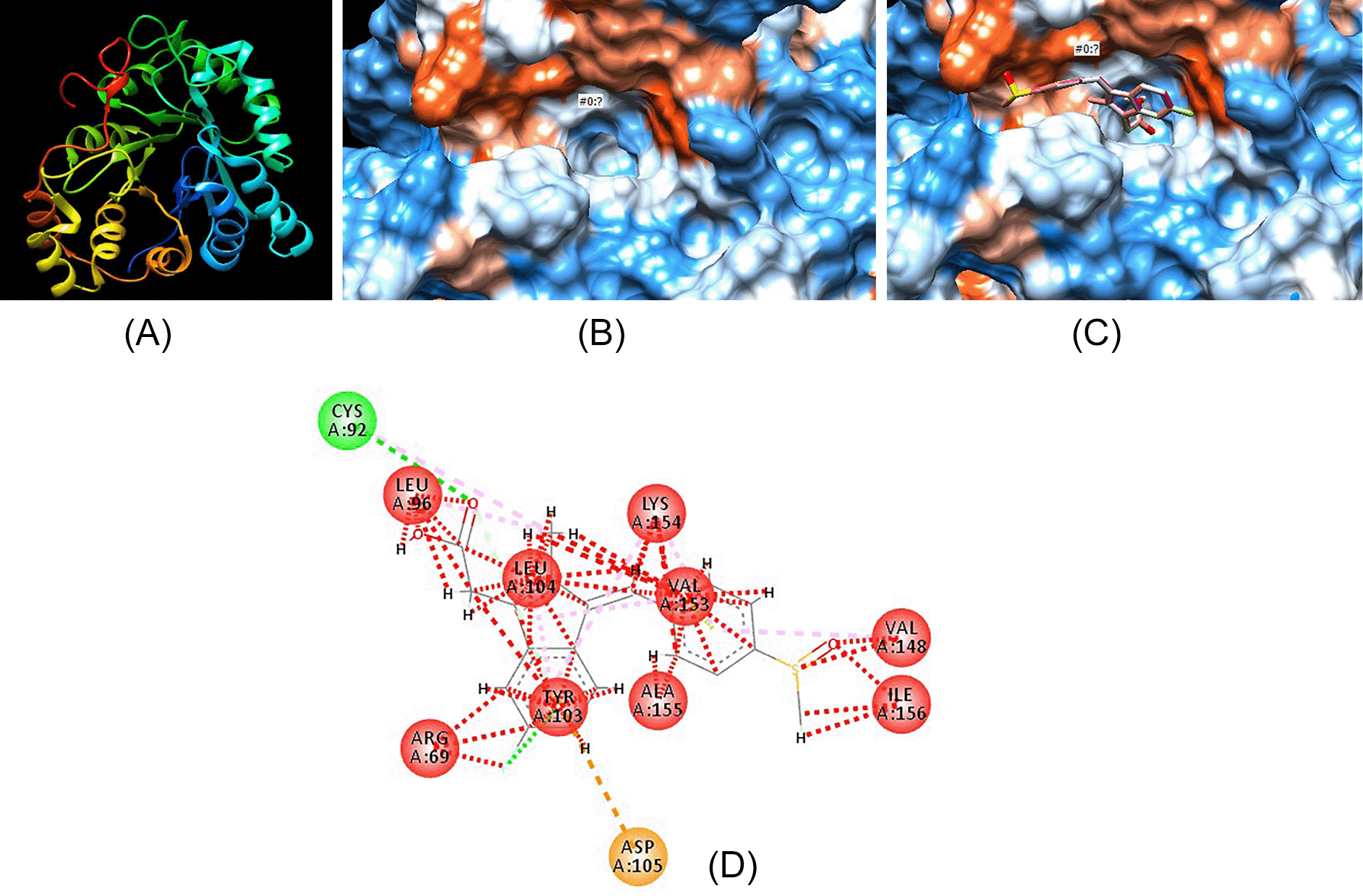
(A) 3D peptide chains of the enzyme aldose reductase. (B) Hydrophobic surface of active binding pocket of aldose reductase. (C) Sulindac fitting in the hydrophobic binding pocket of aldose reductase. (D) Bond interaction between sulindac atoms and amino acids present on the walls of active binding pocket of aldose reductase.
Figure 4 shows the interaction of aldose reductase enzyme with luteolin and its highest binding analogue, ZINC000004349582. Figures 4A and 4C show how luteolin and ZINC000004349582 fits into the binding pocket, while Figures 4B and 4D show the bond interaction between enzyme proteins and luteolin (Figure 4B) and ZINC000004349582 (Figure 4D).
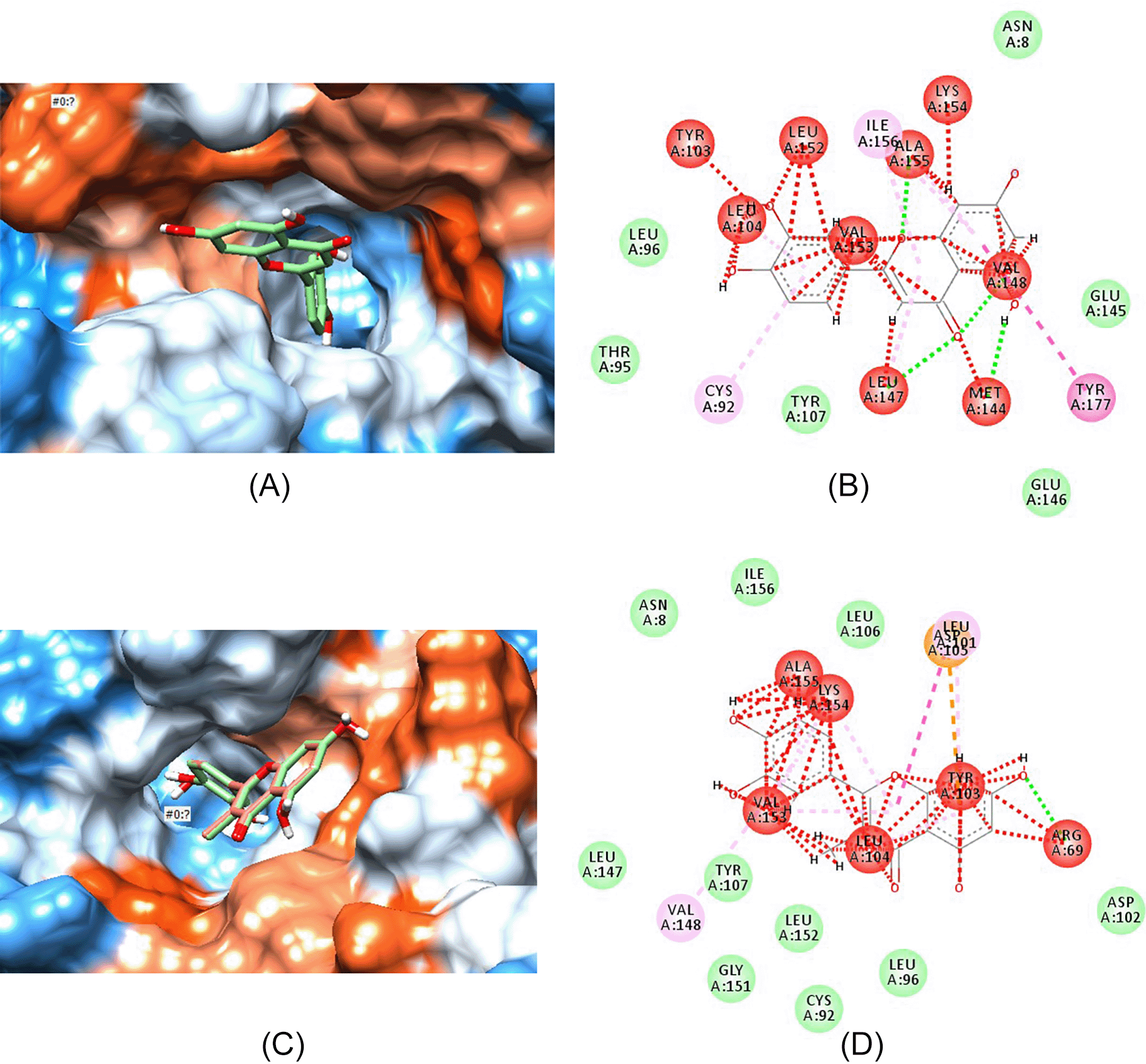
(A) Luteolin fitting in the hydrophobic binding pocket of aldose reductase. (B) Bond interaction between luteolin atoms and amino acids present on the walls of active binding pocket of aldose reductase. (C) ZINC000004349582 fitting in the hydrophobic binding pocket of aldose reductase. (D) Bond interaction between atoms in compound ZINC000004349582 and amino acids present on the walls of active binding pocket of aldose reductase.
Figure 5 shows the interaction of quercetin and its highest binding analogue, ZINC000000057845, with aldose reductase enzyme. Figures 5A and 5C show how quercetin and ZINC000000057845 fits into the binding pocket, while Figures 5B and 5D show bond interactions between enzyme proteins and quercetin (Figure 5B) and ZINC000000057845 (Figure 5D).
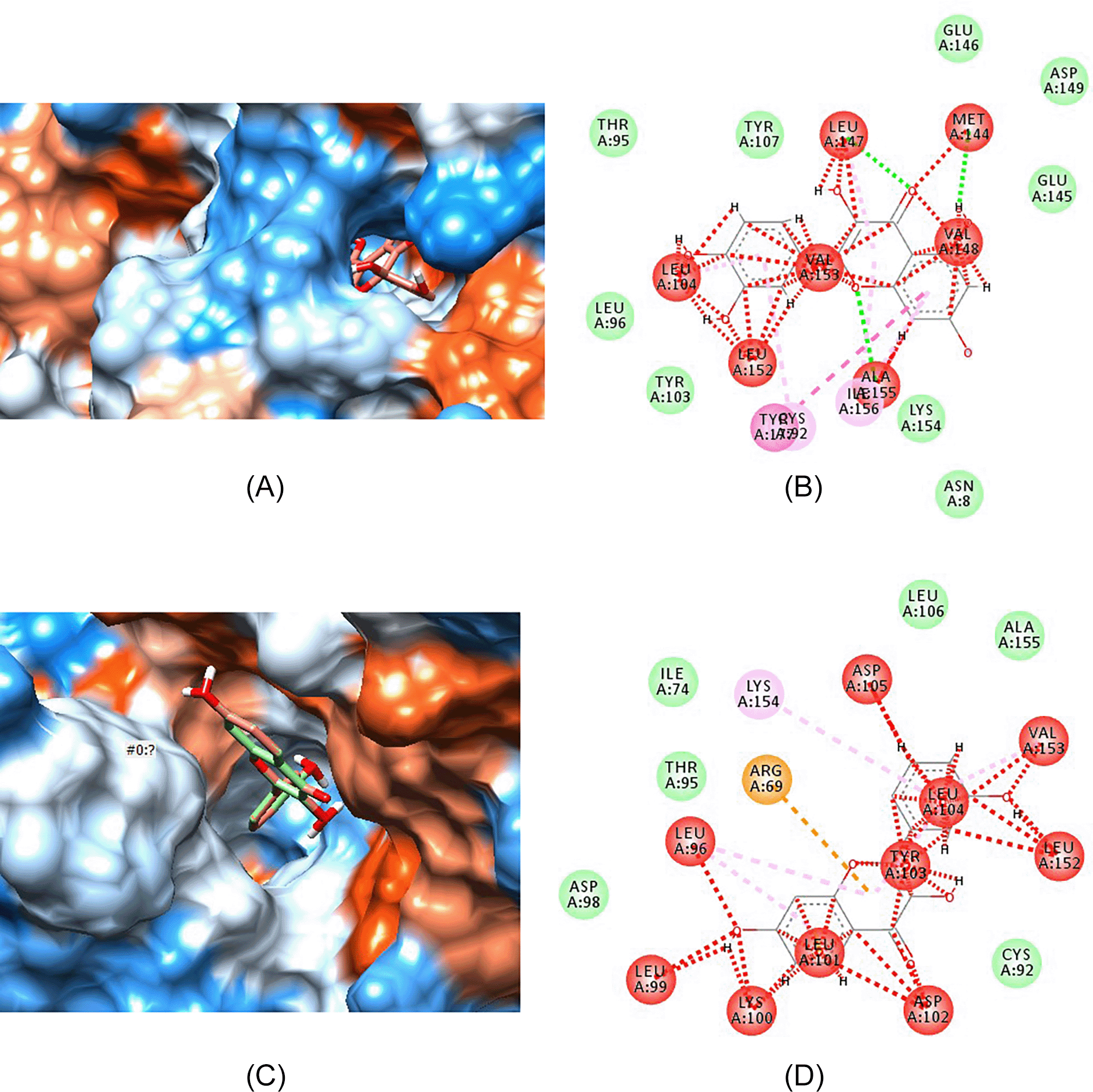
(A) Quercetin fitting in the hydrophobic binding pocket of aldose reductase. (B) Bond interaction between quercetin atoms and amino acids present on the walls of active binding pocket of aldose reductase. (C) ZINC000000057845 fitting in the hydrophobic binding pocket of aldose reductase. (D) Bond interaction between atoms in compound ZINC000000057845 and amino acids present on the walls of active binding pocket of aldose reductase.
Several plants have been shown to specifically inhibit aldose reductase enzyme, which is implicated in the development of diabetic complications (Saraswat et al., 2008). Further, synthetic derivatives have also been formulated but remain unsuccessful when it comes to clinical trials (Singh Grewal et al., 2015). As such, none are yet approved as medication for the management of either retinopathy, nephropathy or neuropathy. Aldose reductase inhibitors (ARIs) have been shown to bind at a separate site from that of NADPH and glucose, which is highly hydrophobic as presented in Figure 3B. Studies on structural activity relationships show that ARIs need to have a primary lipophilic moiety, often an aromatic ring and a thiocarbonyl or carbonyl group that is located 2.8 to 3.8A from the center of the primary group. Acetylsalicylic acid and sulindac (Figure 2B) conform to such characteristics but higher concentrations than the therapeutic range are required to inhibit the enzyme. The addition of a second lipophilic group has been shown to increase inhibitory activity as seen in compounds such as quercetin (Figure 2C) even at micro molar concentrations (in general flavonoids) (Kawanishi et al., 2003). The phytochemicals analyzed in this study were thus from the flavonoid group, luteolin and quercetin. Both phytochemicals were predicted to have 100% probability of binding the enzyme as presented in Table 1.
Comparatively, both luteolin and quercetin compounds were predicted to have a slightly stronger binding affinity (-9.7) for aldose reductase enzyme than sulindac (-9.6) as presented in Tables 2 and 3. A docking score of below -8.0 is generally taken as better binding strength as depicted by both compounds. Eight out of the 20 analogues of luteolin had docking below both sulindac and the parent phytochemical, with ZINC000004349582 having the strongest predicted binding strength (-10.5) as shown in Table 2. Moreover, all compounds were 99.8% or above similar to luteolin. Analysis of quercetin analogues showed that 14 out of the 20 selected ZINC compounds had better binding affinity than both parent phytochemical and the comparator. In particular, the compounds ZINC000000039111, ZINC000575623588, and ZINC000000057845 had the highest docking scores (-10.6) even slightly higher than those of luteolin. Furthermore, their similarity index was above 99.7% as seen in Table 3.
Luteolin, quercetin, sulindac, ZINC000004349582 and ZINC000000057845 all fit within the hydrophobic binding pocket of aldose reductase as shown in Figures 4A, 5A, 3C, 4C and 5C, respectively. The hydrophobic pocket structural analysis of the complexes formed between the ligands and the enzyme showed that the benzaldehyde-carbonyl moiety in sulindac (Figures 3C and 3D) interacts with the amino acids valine-148 and isoleucine-156 of the peptide chain. Conversely, the chromene ring in both luteolin (Figure 2) and quercetin (Figure 4) has the keto substituent bearing the carbonyl that interacts with methionine-144 (Figures 4B and 5B). The valine-148 that interacted with carbonyl moiety in sulindac (Figure 3D) interacts with the benzyl portion of chromene ring in both phytochemicals (Figures 4B and 5B). The substituted phenyl ring serves as the second lipophilic group that is proposed to increase inhibitory activity seen in flavonoids. However, structural analysis of luteolin and quercetin best analogues, showed that the chromene keto group interacts with leucine-104 (Figure 4D) and aspartate-102 (Figure 5D), respectively.
More interactions of the analogues with aldose reductase are seen with the substituted phenyl ring attached to the chromene ring. In particular, the hydroxyl groups and the hydrogen atoms in both analogues may interact with any of the following amino acids: leucine-104, leucine-152, valine-153, tyrosine-103, alanine-155, and lysine-154. In summary, peptide amino acids running between the 100 and 150 position form active site of such molecules and may interact with them.
Pharmacokinetic profile prediction showed that luteolin (Table 2), quercetin (Table 3) and the comparator (sulindac) did not violate the Lipinski rule of five and had a high GI absorption probably due to the ring systems that cancel out the hydrophilic nature of the attached group, thus making the molecules relatively neutral. Additionally, both phytochemicals were predicted to neither cross the blood brain barrier nor be a substrate of p-gp (also known as MDR1, ABCB1 or CD243) as presented in Tables 2 and 3, respectively. The p-gp protein functions to efflux drugs into the intestinal lumen, reducing absorption and bioavailability (Amin, 2013). Though both phytochemicals were predicted to not inhibit CYP2C19 and CYP2C9, they were however inhibitors of CYP1A2, CYP2D6 and CYP3A4. On the contrary, the comparator inhibited all cytochrome enzymes analyzed except CYP1A2 and CYP2D6. All the eight and 14 analogues of luteolin and quercetin had similar pharmacokinetic profiles as their parent phytochemical compounds due to high structural similarity. As such, the ZINC molecules could induce potential drug-drug interaction via the prevention of other drug metabolism.
On toxicological analysis, luteolin was predicted to be safer, falling under toxicology class 5 (LD50=3,919 mg/kg), while both quercetin and sulindac were relatively unsafe, falling under toxicology class 3 (LD50=159 mg/kg and LD=264 mg/kg, respectively) (Gadaleta et al., 2019). Two out of the eight luteolin analogues also fell under toxicology class 3, while the rest were under class 5 with similar predicted LD50 values, as shown in Table 2. In comparison, five quercetin analogues had LD50 of 159 mg/kg, while another five had LD50=3,919 mg/kg. Two of the remaining four quercetin analogues had LD50=4,000 mg/kg, while the other two had a LD50 of 5,000 mg/kg, as presented in Table 3. The luteolin analog with the strongest binding affinity had an LD50 value of 3,919 mg/kg, while for quercetin the best docking compounds, compound ZINC000000039111 and ZINC000575623588, had an LD50 of 159 mg/kg, while ZINC000000057845 had an LD50 of 4,000 mg/kg.
In conclusion, luteolin and quercetin had better docking scores and, thus, higher binding strength compared with sulindac. A total of eight out of the 20 luteolin analogues had docking scores more negative than parent phytochemical compound, and 14 out of the 20 quercetin analogues had docking scores more negative than parent phytochemical compounds. Luteolin analogue (ZINC000004349582) and quercetin analogues (ZINC000000039111, ZINC000575623588, and ZINC000000057845) had the most negative scores (-10.5 and -10.6, respectively) and thus the strongest predicted binding affinity. Both phytochemicals and the eight and 14 analogues had similar pharmacokinetic profiles, with all obeying the Lipinski rule, having a high GI absorption, neither crossing the blood brain barrier nor being acted upon by p-gp and were inhibitors of CYP1A2, CYP2D6 and CYP3A4. Luteolin was predicted to be relatively safer than both quercetin and sulindac. Most analogues of luteolin were generally safer, while the majority of quercetin analogues had greater LD50 values compared with luteolin.
Harvard Dataverse: VIRTUAL SCREENING FOR CHEMICAL ANALOGUES SIMILAR TO PHYTOCHEMICALS THAT INHIBIT ALDOSE REDUCTASE IN THE DEVELOPMENT OF DIABETIC MICROVASCULAR COMPLICATIONS. https://doi.org/10.7910/DVN/3Y2SSD (Otieno, 2022).
This project contains the following underlying data:
- New folder.rar (Contains data on: how the phytocompounds, their analogues and sulindac were docked to the enzyme aldose reductase and the complexes formed. The file types contained in the zipped file are CONF, .PDB, .PDBQT, .MOL2, AND.SDF. All file types can be opened with Chimera software)
- Project analysis.xlsx (Docking score results, pharmacokinetic and toxicological analysis of zinc analogues)
- sulindac then luteolin then quercetin ADME.tab (Pharmacokinetic analysis from SwissADME for parent phytocompounds and comparator)
- swissadme of analogues (1).xlsx (Pharmacokinetic analysis from SwissADME for zinc analogues of parent phytocompounds)
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
We presented this research at the 2nd Global Webinar on Diabetes and Endocrinology conference held on November 28-29, 2022.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
No source data required
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: In silico study, synthetic chemistry
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Drug design
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 21 Mar 23 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)