Keywords
5-aminolevulinic acid, 5-aminolevulinic acid dehydratase, hemB, hemA, hemL, rhtA
This article is included in the Japan Institutional Gateway gateway.
Microbial production of 5-aminolevulinic acid (ALA) attracts attention due to a wide range of biotechnological and medical applications of ALA, including cancer treatment and diagnosis. Various genetic engineering approaches have been employed to improve ALA production in bacterial hosts such as Escherichia coli possessing the C5 pathway. Glutamyl-tRNA reductase (GluTR) encoded by hemA, glutamate-1-semialdehyde aminotransferase (GSA-AT) encoded by hemL, and ALA dehydratase (ALAD) encoded by hemB play important roles in ALA metabolism including the C5 pathway. Attenuation of the intercellular ALAD activity, which condensates 2 molecules of ALA to synthesize porphobilinogen (PBG), has been employed by various measures. However, a mutation approach by substituting catalytically important residues in ALAD encoded by hemB has never been attempted. The aim of this study is to assess the impact of hemB mutations on the ALA production in E. coli.
In this study, the authors mutated the amino acid residues potentially related to the enzymatic activity of E. coli ALAD by referring to a mutation experiment of human ALAD. The authors created five types of mutated hemB genes, introduced these genes to the hemB-deleted mutant strain of E. coli, and assessed the impact of the ALAD mutations on ALA production. In addition, hemA, hemL, and rhtA encoding an ALA exporter were introduced to the E. coli possessing a mutated hemB.
The authors revealed that the mutations of ALAD employed in this study did not significantly enhance ALA production. Overexpression of hemA, hemL, and rhtA substantially increased ALA production in any E. coli strain possessing mutated hemB, while a difference in ALA production of the strain could be rather attributed to its growth behaviour than ALAD inactivation.
This study provides an important piece of information to design the bioprocess of ALA production using E. coli engineered through the C5 pathway.
5-aminolevulinic acid, 5-aminolevulinic acid dehydratase, hemB, hemA, hemL, rhtA
5-aminolevulinic acid (ALA) is an intermediate in the synthetic pathway of porphyrin derivatives indispensable to life such as heme, vitamin B12 and chlorophyll, and thus found in a wide range of prokaryotic and eukaryotic organisms.1 ALA has been useful compounds or agents of very considerable interest because it is applicable to various biotechnologies including diagnosis and treatment of cancer, biodegradable herbicide, and growth promotion factor for plants.2
Industrial production of ALA was conventionally performed using a photosynthetic bacterium Rhodobacter sphaeroides possessing the C4 pathway in which ALA synthase (EC 2.3.1.37) produces ALA from glycine and succinyl-CoA.3 The mutation and metabolic engineering approaches of R. sphaeroides have successfully demonstrated a significant improvement in ALA productivity.4 Another option is to use bacteria (including Escherichia coli) that possess another pathway of ALA-synthesis referred to as the C5 pathway, which originates with glutamate (Glu) and subsequently glutamyl-tRNA, glutamate-1-semialdehyde leading to ALA (Figure 1).5–7 The enzymes that specify the C5 pathway to produce the precursors of ALA accumulation are glutamyl-tRNA reductase (GluTR, EC 1.2.1.70) encoded by hemA and glutamate-1-semialdehyde aminotransferase (GSA-AT, EC 5.4.3.8) encoded by hemL. In the downstream pathway of the C5 pathway, ALA dehydratase (ALAD, EC 4.2.1.24) encoded by hemB condensates two molecules of ALA to synthesize porphobilinogen (PBG), which is converted to various types of tetrapyrrole including heme. Membrane transporter RhtA encoded by rhtA serves as an exporter of ALA.
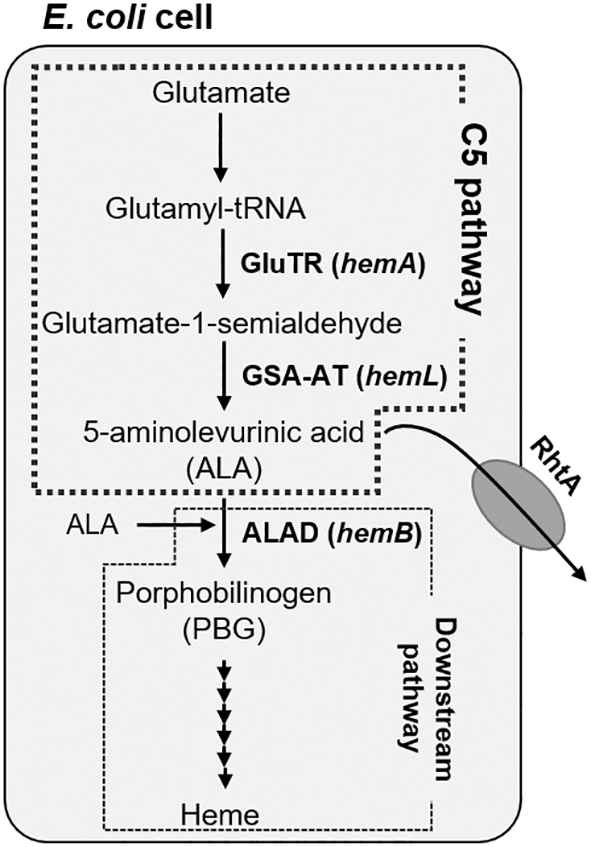
The genes hemA encoding glutamyl-tRNA reductase (GluTR), hemL encoding glutamate-1-semialdehyde aminotransferase (GSA-AT), hemB encoding ALA dehydratase (ALAD), and RhtA-encoding gene were engineered in this study.
Various types of genetic engineering approaches have been examined to enhance ALA production through the C5 pathway. Kang et al., obtained a high ALA accumulation in E. coli through the synergetic effect of the endogenous hemL and heterologous hemAM that was mutated to stabilize the encoding GluTR structurally.8 They also found that overexpression of rhtA increased ALA production. Inhibiting the conversion of ALA to porphyrin derivatives is also a well-known strategy to improve ALA production. Since the ALDA encoded by the hemB gene plays a crucial role in synthesizing porphyrin derivatives essential to the cellular growth, an efficient inhibitory node can be arranged on the downstream metabolic pathways not by completely knocking-out hemB but by lowering the ALAD enzyme activity (Figure 1). Zhang et al., verified inactivation of the ALAD enzyme activity in Corynebacterium glutamicum as reduced to 47% and 69% in contrast with the enhancement of ALA accumulation as 1.34 and 1.19 times, respectively, by replacing the intrinsic ribosome-binding site (RBS) of hemB to relatively weak RBS along with overexpression of hemA, hemL and rhtA.9 Attenuation of the hemB expression was also achieved by some other approaches such as promoter exchanging,10 CRISPRi,11 and antisense RNA expression.12 Although researchers have examined various approaches to downregulation of the transcriptional or translational levels of hemB for enhancement of ALA production, the authors have first attempted the mutation approach to lower the enzymatic activity of ALAD.
It has been known that the human ALAD forms quaternary homo-oligomers (four types of morpheein forms) of high-activity octamer, transient hugging dimer, transient detached dimer, and low-activity hexamer.13 In rare cases, heteromeric oligomers were found in the human ALAD of patients with ALAD porphyria with more than one genetic aberration.13 The heteromeric oligomers comprise the wild-type K59 or N59 and the mutant F12L. The F12L variant forms a stable homohexamer and possesses a significantly low enzymatic activity, whereas the wild type K59 and N59 variants form a stable homooctamer (hexameric morpheein concentration is 0% and 3%, respectively) and possess a high enzymatic activity. Jaffe and Stith demonstrated that porphyria-associated mutation in the human ALAD causes the morpheein equilibrium to be shifted toward the inactive hexamer.13 Inspired by the study, the authors considered that some mutations in the E. coli ALAD also have a certain possibility of the morpheein shift toward the hexameric assembly for leading to enzymatic inactivation and an increase in ALA accumulation of the genetically engineered E. coli.
In the present study, the specific amino acid residues on E. coli ALAD was first identified to correspond to the enzymatically important residues on the human ALAD, and then inactive residues were substituted for the specific residues. Subsequently, the impacts of these mutations on ALA accumulation in the E. coli cells were analysed. Furthermore, the synergetic effects of these ALAD mutations and overexpression of RhtA, GluTR (encoded by hemA) and GSA-AT (encoded by hemL) were also examined.
E. coli strain W3110 hemB (mutant with hemB deletion, kanamycin resistance) was obtained from National BioResource Project (National Institute of Genetics (NIG), Shizuoka, Japan). Molecular cloning was performed using E. coli strain JM109. E. coli was cultured at 37°C in the lysogeny broth (LB) medium containing appropriate combinations of 50 μg/mL kanamycin sulphate (FUJIFILM Wako Pure Chemical Co., Catalogue number: 113-00343), 50 μg/mL chloramphenicol (FUJIFILM Wako Pure Chemical Co., Catalogue number: 034-10572), 100 μg/mL spectinomycin dihydrochloride pentahydrate (FUJIFILM Wako Pure Chemical Co., Catalogue number: 152067) or 150 μg/mL ampicillin sodium (FUJIFILM Wako Pure Chemical Co., Catalogue number: 012-23303) for selection. Isopropyl-β-D-thiogalactopyranoside (IPTG, final concentration: 0.1 mM, TaKaRa Bio Inc., Catalogue number: 9030) was added to induce the expression of the genes under the control of the lac promoter.
Protein structures of ALAD derived from Homo sapience (PDB 1E51) and E. coli (PDB 1B4E) were obtained from the Protein Data Bank (PDB). Alignment of the protein structures was carried out using PyMoL (RRID:SCR_000305) (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.).
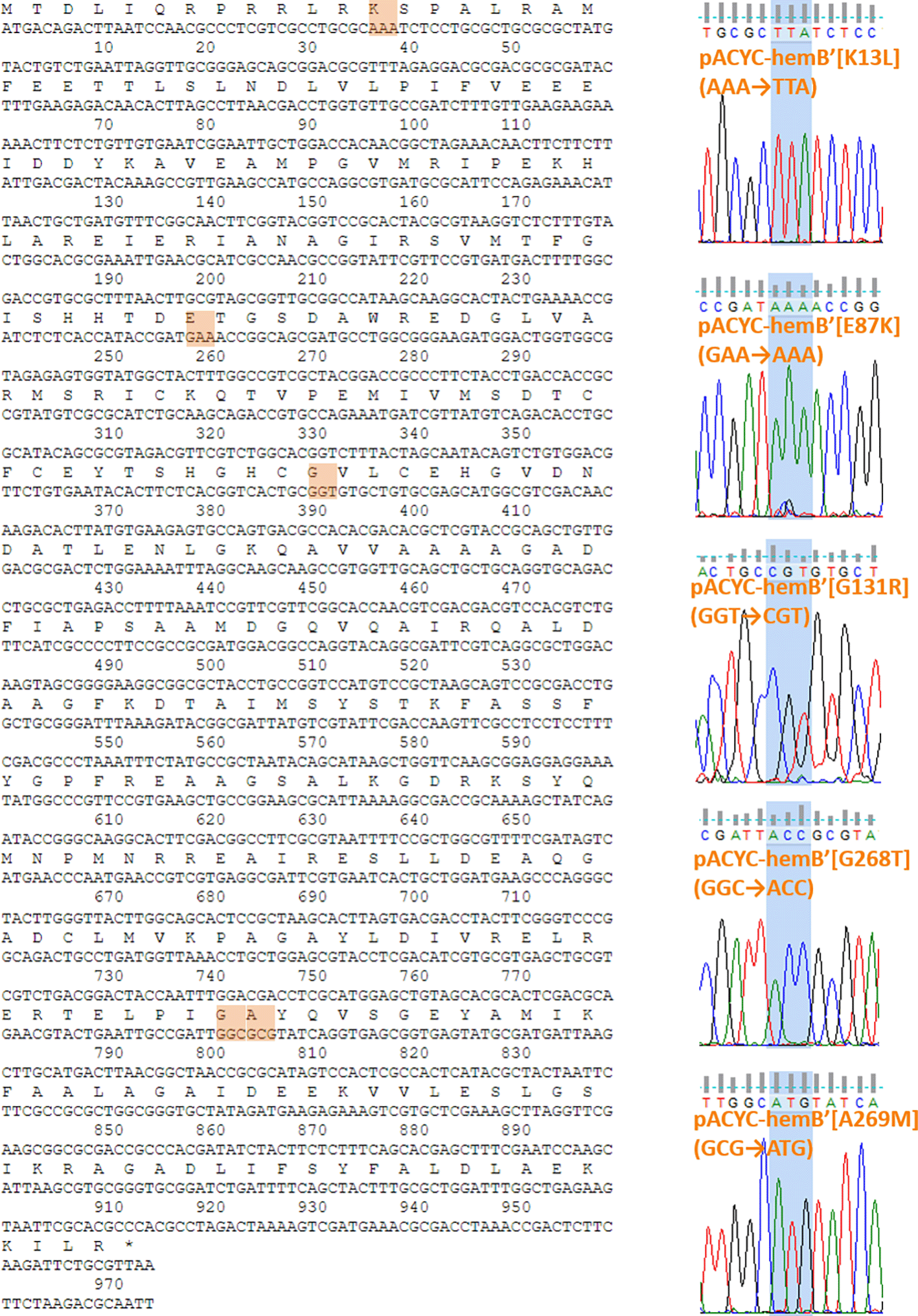
Primers, plasmids, and E. coli strains used in this study are listed in Tables 1, 2, and 3, respectively. pACYCDuet-1 (carrying the P15A replicon origin, ~10 copies/cell) was purchased from Sigma-Aldrich Co. (Catalogue number: 71147, St. Louis, MO, USA). A partial fragment of pACYCDuet-1 without lac-promoter was amplified by PCR using primers of pACYCDuet_F and pACYCDuet_R and PrimeSTAR Max DNA polymerase (TaKaRa Bio Inc., Shiga, Japan, Catalogue number: R045A). PCR amplification experiments were performed using the PCR machines (Takara Bio Inc., model TP350 and Blue-Ray Biotech Co., TurboCycler, model TCST-9612) with the thermal cycling condition unless otherwise noted; after the initial denaturation step at 98°C for 1 min, 30 cycles of the denaturation step at 98°C for 10 sec, annealing step at 50~68°C for 5 or 15 sec, and extension step at 72°C for 1 min/kbp, followed by an additional extension step at 72°C for 4 min. The hemB along with native hemB promoter was amplified by PCR using primers of hemB promoter+hemB_F and hemB promoter+hemB_R with the extracted E. coli genome as a template. DNA concentrations were measured using NanoDrop 1000 (Thermo Fisher Scientific, Waltham, MA, USA). These fragments were assembled by In-Fusion HD Cloning Kit (TaKaRa Bio Inc., Catalogue number: 639634) at 50°C for 15 min. The assembled samples were introduced into E. coli strain JM109 by heat shock method (42°C, 45 sec). The colonies were cultivated in LB agar (1.5%) medium containing 50 μg/mL chloramphenicol. The assembly of the plasmid was confirmed by restriction enzyme treatment, followed by Sanger sequencing. The resulting plasmid were designed as pACYC-hemB. Additionally, five types of plasmids containing mutated hemB (namely hemB’[K13L], hemB’[E87K], hemB’[G131R], hemB’[G268T], hemB’[A269M]) were constructed by inverse PCR using pACYC-hemB as a template and one of the following primer sets, hemB_K13L_F and hemB_K13L_R, hemB_E87K_F and hemB_E87K_R, hemB_G131R_F and hemB_G131R_R, hemB_G268T_F and hemB_G268T_R, and hemB_A269M_F and hemB_A269M_R. The amplified fragments were assembled as described above. The resulting plasmids were designed as pACYC-hemB’[K13L], pACYC-hemB’[E87K], pACYC-hemB’[G131R], pACYC-hemB’[G268T], and pACYC-hemB’[A269M], respectively.
F, forward; R, reverse.
pCL1920 (carrying the pSC101 replicon origin, ~5 copies/cell) was purchased from National BioResource Project (NIG). A partial fragment of pCL1920 by PCR using a primer sent of pCL1920_F and pCL1920_R. The rhtA was amplified by PCR using primers of rhtA_In_pCL1920_F and rhtA_In_pCL1920_R with the extracted E. coli genomic DNA as a template. The amplified fragments were assembled as described above. The colonies were cultivated in LB agar medium containing 100 μg/mL spectinomycin. The resulting plasmid was designed as pCLS-rhtA.
pMD19 (derived from pUC19 with ~500 copies/cell) was purchased from TaKaRa Bio (Catalogue number: 3271). A partial fragment of pMD19 was amplified by PCR using primers of pMD19_In_hemL_F and pMD19_In_hemL_R. The hemA and hemL, both derived from E. coli, were amplified using primers of hemA_In_pMD19_F and hemA_In_hemL_R, and hemL_In_hemA_F and hemL_In_pMD19_R, respectively. Subsequently, the amplified fragments of hemA and hemL were together assembled with the partial fragment of pMD19 by the method described above using LB agar medium containing 150 μg/mL ampicillin. The resulting plasmid was designed as pMD-hemA-hemL.
Partial fragments of pCLS-rhtA and pMD-hemA-hemL were amplified by PCR using primer sets of pCLS-rhtA_F and pCLS-rhtA_R, and hemA-hemL_F and hemA-hemL_R, respectively. These fragments were assembled by the method described above. The resulting plasmid was designed as pCLS-rhtA-hemA-hemL containing rhtA, hemA and hemL.
The amount of ALA production was measured by colorimetric quantification using a modified Ehrlich’s reagent prepared by mixing 1 g of dimethylaminobenzaldehyde (DMAB, FUJIFILM Wako Pure Chemical Co., Catalogue number: 047-18042), 30 mL of acetic acid (FUJIFILM Wako Pure Chemical Co., Catalogue number: 017-00256), 8 ml of 60% (v/v) perchloric acid (Kanto Chemical Co. Inc., Catalogue number: 32059-1B), followed by addition of acetic acid to make the total volume 50 mL.14 The supernatant of culture (100 μL) was added to 100 μL of the mixture (1:99 vol. ratio) of acetylacetone and acetate buffer (prepared by mixing 91 mL of 0.1 M acetic acid and 109 mL of 0.1 M sodium acetate solution, pH 4.7), and incubated at 100°C for 15 min. The mixture was then cooled down on ice. The modified Ehrlich’s reagent (200 μL) was added to the mixtures and incubated at room temperature for 15 min. Ultrapure water (600 μL) was added, and absorbance at 553 nm was measured using a spectrophotometer UV-1900 (Shimadzu Co., Kyoto, Japan).
Figure 2 illustrates both the structures of the E. coli ALAD (in orange) and human ALAD (in blue) resemble each other. Figure 2 also demonstrates that the K13, E87, G131, G268 and A269 residues on the E. coli ALAD occupy the positions corresponding to the F12, E89, G133, A274 and V275 residues on the human ALAD. The conformational characteristics of these residues guided us to design mutations of the K13L, E87K, G131R, G268T and A269M variants in the E. coli ALAD following the inactive F12L, E89K, G133R, A274T and V275M variants in the human ALAD.13 The authors applied an inverse PCR method to introduce mutations to hemB on the pACYC-based plasmids (pACYC-hemB’[K13L], pACYC-hemB’[E87K], pACYC-hemB’[G131R], pACYC-hemB’[G268T] and pACYC-hemB’[A269M] listed in Table 2) to express these mutated E. coli ALAD. Successful mutations of hemB were confirmed by Sanger sequencing (Figure 3).
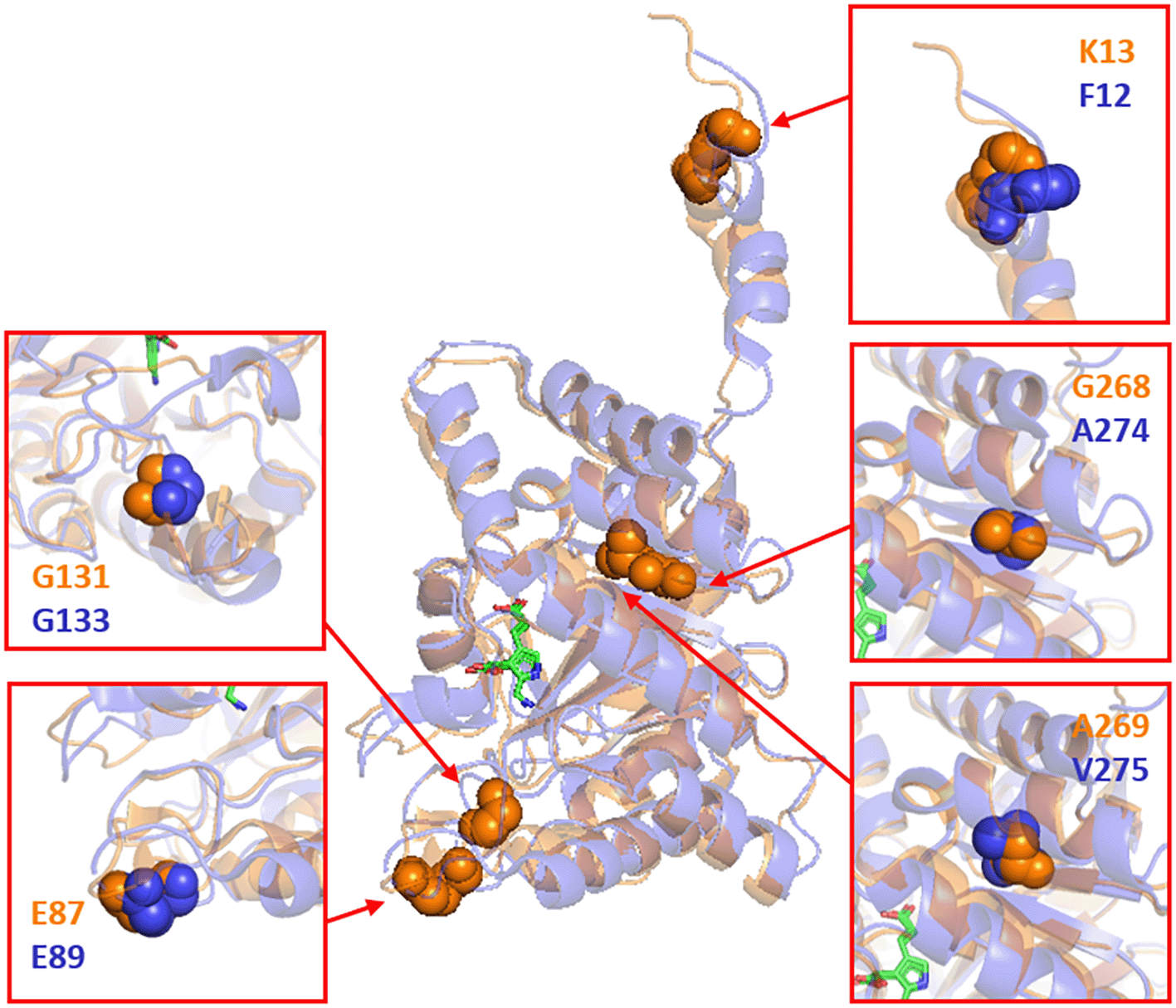
K13, E87, G131, G268, and A269 on the E. coli ALAD structurally correspond to F12, E89, G133, A274, and AV275 on the H. sapience ALAD, respectively. Porphobilinogen (PGB) products are shown in green wire forms. The figure was prepared using PyMoL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.).
In the present study, the mutant E. coli strain W3110 hemB with hemB deletion (hereinafter referred to as strain ∆B, Table 3) was used as a basic strain to assess the impacts of ALAD mutations. The strain ∆B with a lack of ALAD activity can grow only in the LB medium containing supplementation of 0.2% glucose as well (Figure 4A).15 The strain ∆B_B was established by introducing the intrinsic hemB in the strain ∆B and can grow without supplementation of 0.2% glucose (Figure 4), suggesting that ALAD activity was essential to support the E. coli growth in the absence of glucose. When one of the five types of mutated hemB was introduced to the strain ∆B, every strain (namely, the strains ∆B_B’[K13L], ∆B_B’[E87K], ∆B_B’[G131R], ∆B_B’[G268T], or ∆B_B’[A269M], Table 3) showed steady growth even in the LB medium without supplementation of glucose (Figure 4). This result indicates that the above-mentioned five types of the mutated hemB expressed the enzymatically active ALAD in E. coli. Figure 4B showed the comparison of the growth of these strains. The mutant strain harbouring pACYC-hemB’[G131R] exhibited an obviously low growth rate, while the molecular mechanism underlying the growth inhibition remains unclear.
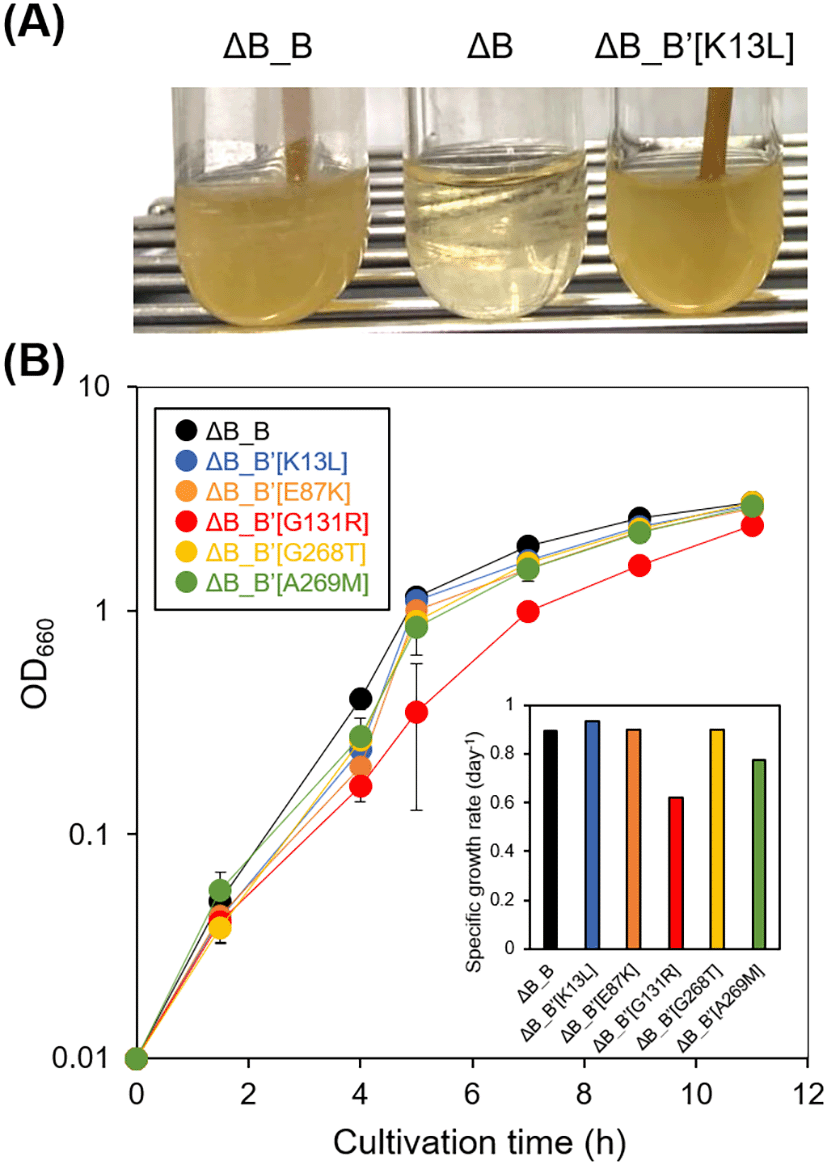
(A) Culture of E. coli strains ∆B, ∆B_B, and ∆B_B’[K13L] in the lysogeny broth (LB) medium after overnight incubation. (B) Growth curves of E. coli strains ∆B_B, ∆B_B’[K13L], ∆B_B’[E87K], ∆B_B’[G131R], ∆B_B’[G268T], and ∆B_B’[A269M] (inset: specific growth rates of each strain).
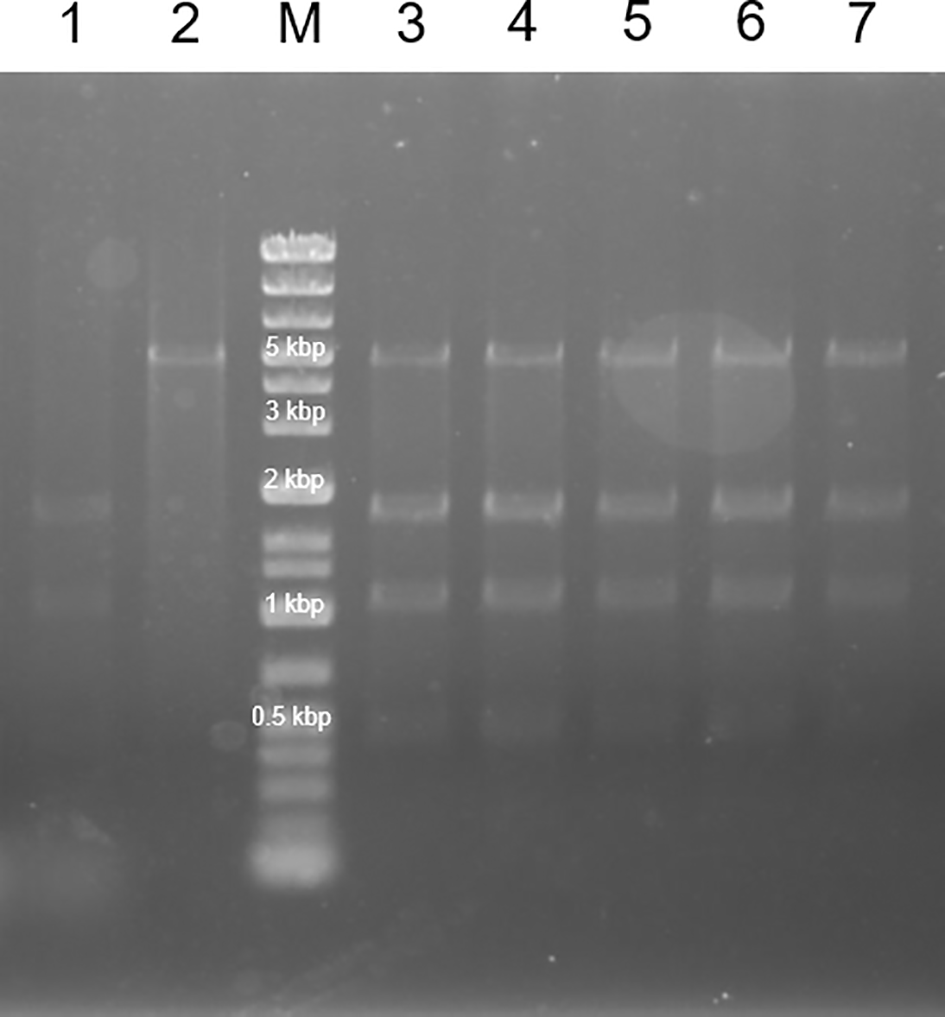
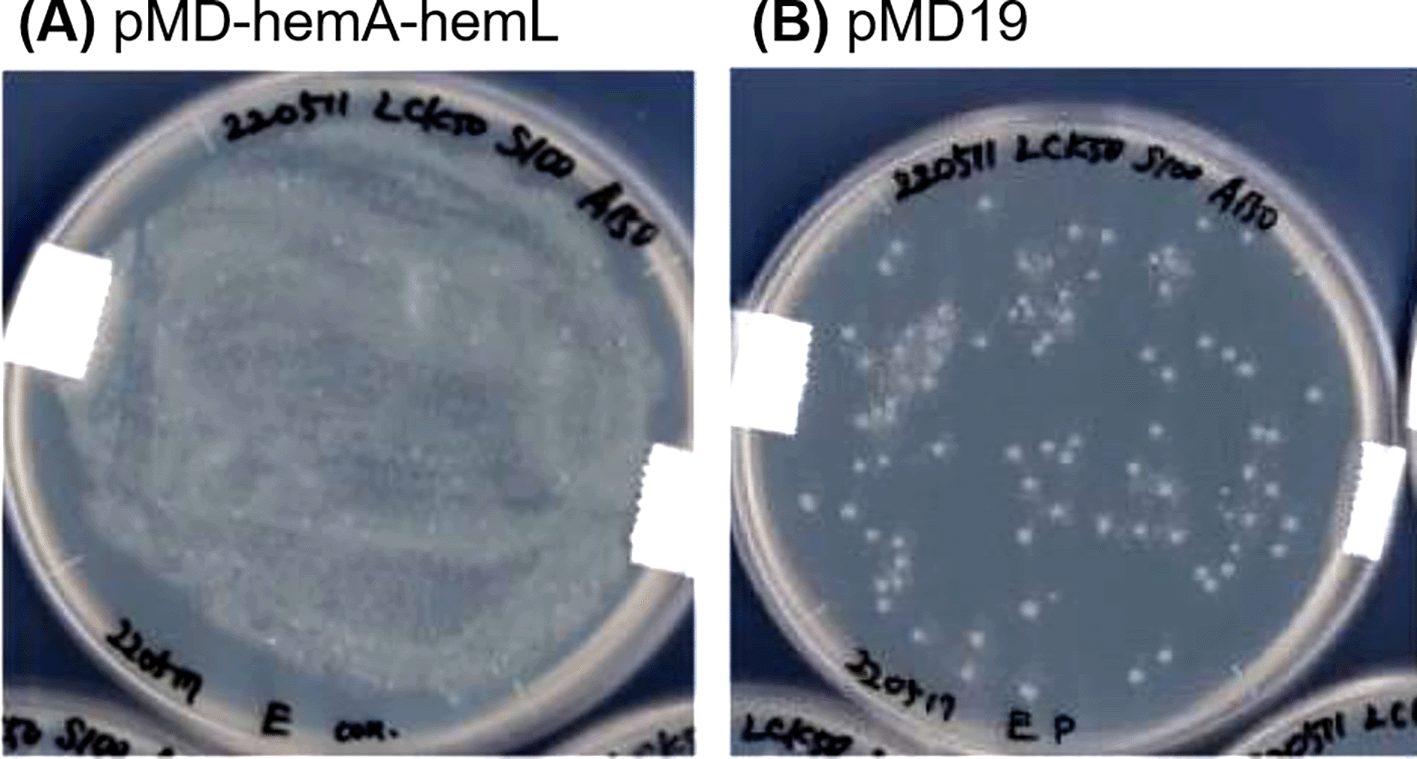
Further engineering of the strains expressing mutated ALAD was performed by introducing the ALA-exporter gene rhtA. We successfully obtained the transformant strains designated as ∆B_B’[K13L]&R, ∆B_B’[E87K]&R, ∆B_B’[G131R]&R, ∆B_B’[G268T]&R and ∆B_B’[A269M]&R (Table 3). The restriction enzyme assay confirmed that the transformed pACYC-based plasmid with mutated hemB and pCLS-rhtA plasmid were maintained in the transformant strains (Figure 5). Moreover, along with the pACYC-based plasmid with mutated hemB and pCLS-rhtA plasmid, the pMD-hemA-hemL plasmid with an ability of highly copying was also introduced to overexpress GluTR and GSA-AT. However, unfortunately, mature colonies did not grow on the LB-agar plates containing antibiotics for selection. Although tiny colonies were observed on the plates (Figure 6A), the cells forming tiny colonies did not grow in the liquid LB medium containing the antibiotics. The introduction of the empty vector pMD19 leaded to the results of the formation of mature colonies (Figure 6B), indicating that the backbone vector pMD19 did not cause the formation of tiny colonies. These results suggest that excess GluTR and GSA-AT expressed from a high copy number of plasmid pMD-hemA-hemL might negatively affect the E. coli growth. To avoid the problem, we replaced the backbone vector pMD19 with pCL1920 having an ability to copy a relatively low number of plasmids for expressing GluTR and GSA-AT. The hemA and hemL encoding GluTR and GSA-AT, respectively, were inserted to the expression cassette of rhtA in pCLS-rhtA. Resultantly, the mature colony of E. coli transformed with the pCLS-rhtA-hemA-hemL plasmid was successfully obtained. The results indicates that the transformant of E. coli was able to harbour mutated hemB (∆B_B’[K13L]&R-A-L, ∆B_B’[E87K]&R-A-L, ∆B_B’[G131R]&R-A-L, ∆B_B’[G268T]&R-A-L and ∆B_B’[A269M]&R-A-L, Table 3). Maintenance of both the pCLS-rhtA-hemA-hemL and pACYC-based plasmid carrying mutated hemB in each transformant clone was confirmed with the restriction enzyme assay (Figure 7). We should emphasize that overexpression of GluTR and GSA-AT in E. coli is a technical challenge involving a risk of their toxic effects on the host cells. As far as we found out, the present study has first achieved to solve the problem by fine-tuning their expression levels with reducing the number of plasmid copies.
Lane M represents the DNA ladder.
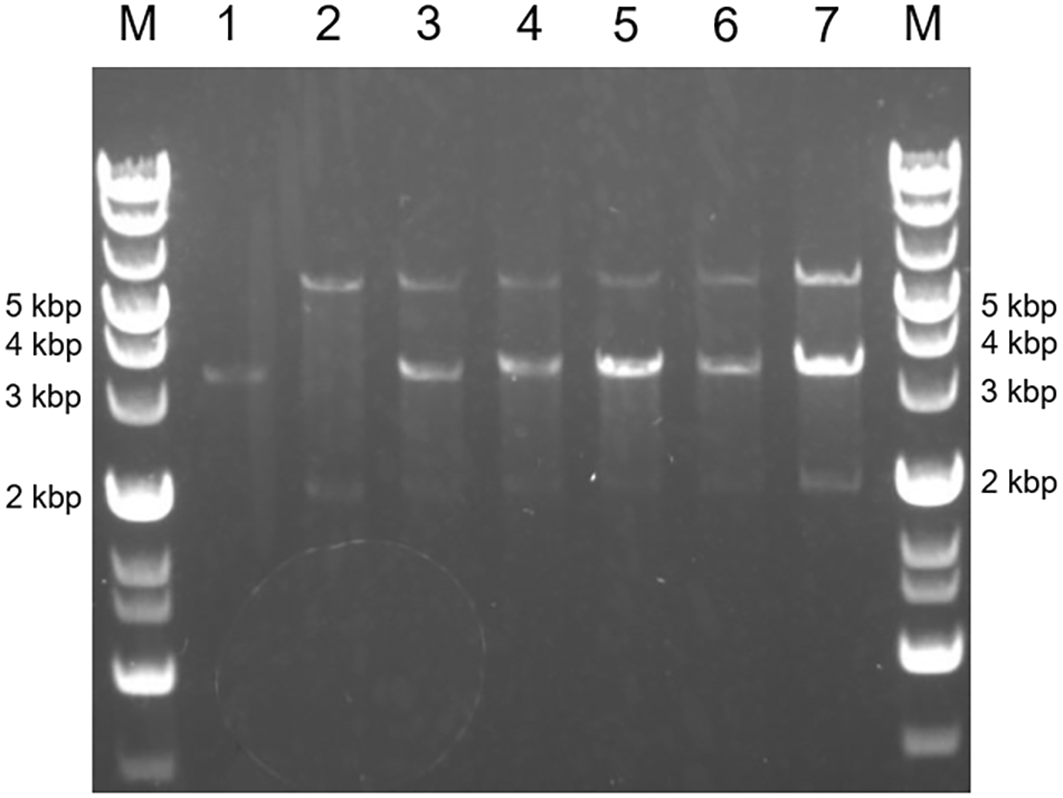
Lane M represent the DNA ladder.
Taking the results mentioned above, the authors constructed three kinds of the series of E. coli transformant strains harbouring the mutated hemB; namely (1) the primary hemB mutation (strains ∆B_B’[mutation]) that simply express the mutated hemB alone, (2) the intermediate hemB mutation (strains ∆B_B’[mutation]&R) that express the mutated hemB along with recombinant rhtA, (3) the triplet hemB mutation (strains ∆B_B’[mutation]&R-A-L) that express the mutated hemB along with recombinant rhtA, hemA and hemL. The ALA production of these transformants were evaluated by the colorimetric measurement. The ALA production of the primary hemB mutation strains was almost uniform under the mutational variation, even though the values were slightly lower than that of the wild type hemB strains (Figure 8A and Table 4). The primary mutation strains did not show any improvement in ALA production of E. coli, suggesting that the primary hemB mutations only slightly decreased the enzymatic activity of ALAD in E. coli, and those impacts on ALA accumulation were hindered by the effects of multiplication of the mutated hemB genes on pACYC-based plasmids (~10 copies/cell). The intermediate hemB mutation strains with overexpression of RhtA from low copy number plasmid marginally improved the ALA production by actively exporting ALA to the supernatant of the culture (Figure 8B and Table 4).
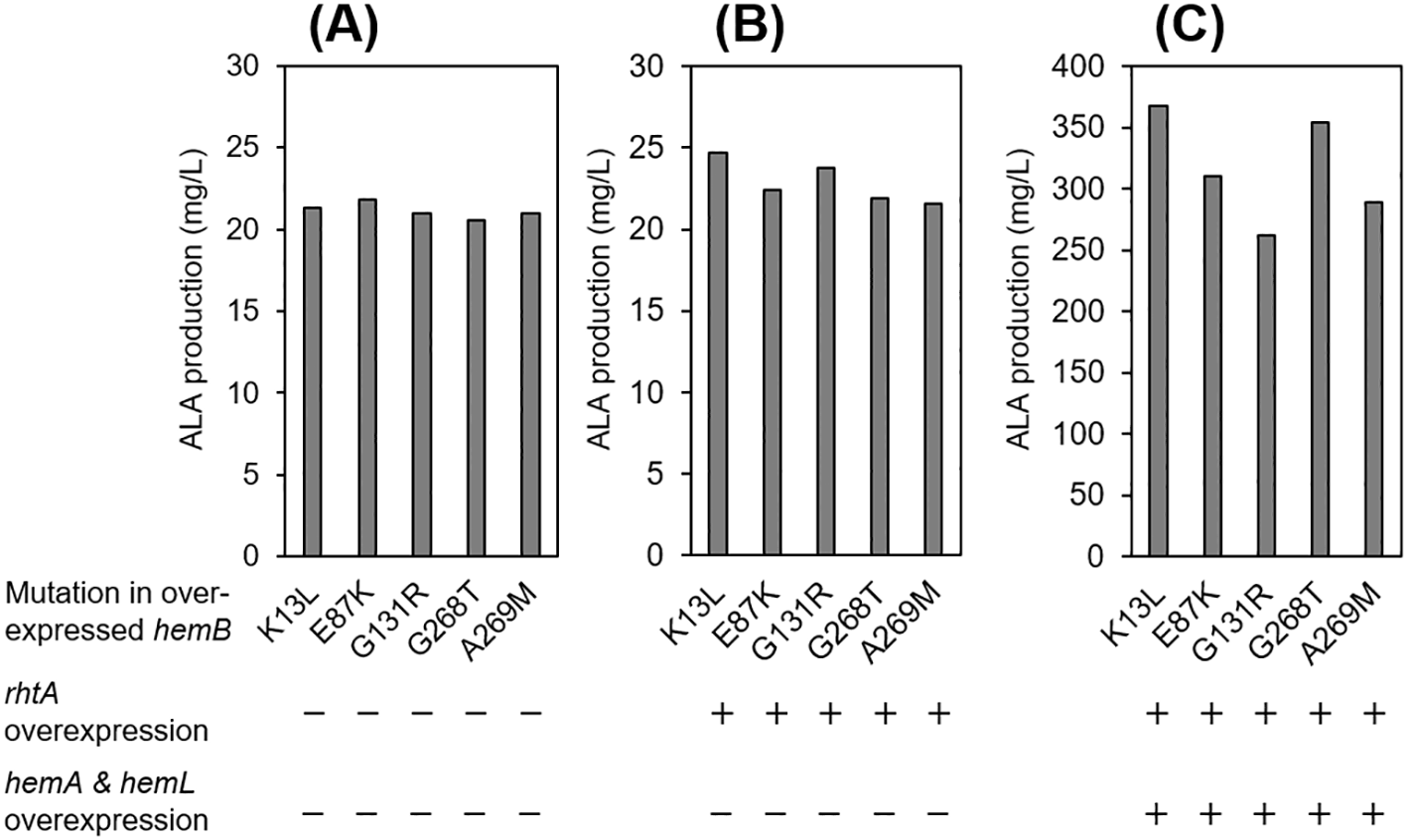
The same data are represented in the form of bar-graph in Figure 4. The unit of the data is mg/L. Data represent the average values of two replicated experiments.
| Type of HemB mutation | rhtA OX1 - | rhtA OX + | rhtA OX + |
|---|---|---|---|
| hemA & hemL OX - | hemA & hemL OX - | hemA & hemL OX + | |
| No mutation | 24.24 | ND2 | ND |
| [K13L] | 21.30 | 24.67 | 368.09 |
| [E87K] | 21.84 | 22.37 | 309.70 |
| [G131R] | 20.96 | 23.76 | 262.39 |
| [G268T] | 20.53 | 21.90 | 354.56 |
| [A269M] | 20.94 | 21.58 | 288.69 |
The triplet hemB mutation strains with further overexpression of GluTR and GSA-AT from low copy number of plasmid carrying hemA and hemL drastically increased the ALA production (Figure 8C and Table 4). The overexpression of GluTR could be expected to lead an enhancement in ALA production because the reaction catalysed by GluTR encoded by hemA is a rate-limiting step for ALA production in the C5-pathway8 and GSA-AT encoded by hemL tightly forms a complex with GluTR.8,16 Clear difference also appeared in ALA production of the transformants with the aid of overexpression of GluTR and GSA-AT depending on the five types of mutated hemB variation. The transformant strains with hemB’[K13L] showed the highest ALA production of 368.1 mg/L, followed those with hemB’[G268T] (354.6 mg/L), hemB’[E87K] (309.7 mg/L), hemB’[A269M] (288.7 mg/L) and hemB’[G131R] (262.4 mg/L). However, we have not been able to attribute the difference appeared in ALA production rigorously to the declining variation in the levels of the ALAD activity caused by the individual mutation because the ALA production depends on both the ALA contents in a unit cell and the biomass production (in other word, growth of cells) of E. coli. The ALA production order of the transformants (hemB’[K13L] was highest, followed by hemB’[G268T], hemB’[E87K], hemB’[A269M] and hemB’[G131R] in Figure 8C and Table 4) is the exact same as the order in the specific growth rates of the corresponding platform strains with mutated hemB (inset figure of Figure 4B). Given these two results; (1) the authors have not discriminated any significant difference of ALA production in the five types of hemB in the platform strains without overexpression of RhtA, GluTR and GSA-AT, (2) the difference of ALA production of the transformant strains with overexpression of RhtA, GluTR and GSA-AT was consistent with the growth behaviour of the platform strains, difference of ALA production shown in Figure 8C could be most likely attributed to not the declining variation of ALAD activity encoded by the mutated hemB, but the different growth of E. coli strains.
Five types of mutations have been introduced on the E. coli ALAD encoded by hemB to induce the accumulation of ALA in E. coli. However, any mutation examined in the present study has not increased ALA production significantly. As far as we found out, the present study has been first performed to assess the impact of mutations of ALAD on ALA production in E. coli. Tuning of the copy number of the plasmid containing hemA and hemL allowed us to obtain the transformant strains expressing rhtA, hemA, hemL, and mutated hemB. Overexpression of RhtA, GluTR (encoded by hemA) and GSA-AT (encoded by hemL) substantially improved ALA production as previously reported. These transformant strains expressing each type of mutated ALAD showed different ALA production, while their different ALA production was most likely attributed to not the declining variation of ALAD activity but the difference in the growth rate of E. coli strains.
Figshare: Gene sequence file (fasta). https://doi.org/10.6084/m9.figshare.24557605.v1. 17
Figshare: Uncropped raw gel images for diagnostic restriction digestion analyses 1. https://doi.org/10.6084/m9.figshare.24556849.v1. 18
Figshare: Uncropped raw gel images for diagnostic restriction digestion analyses 2. https://doi.org/10.6084/m9.figshare.24557308.v1. 19
Figshare: Numeric data for E. coli growth curve. https://doi.org/10.6084/m9.figshare.24556828.v1. 20
Figshare: Output files from PyMoL. https://doi.org/10.6084/m9.figshare.24556768.v1. 21
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: plant physiology
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: soil and environmental microbiology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 23 Feb 24 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)