Keywords
plastome, NGS, coastal zone, phylogenomics
Olive (Olea europaea Linaeus 1753) is one of the valuable fruit trees and very important edible oil plant in the world. The chloroplast (cp) genome of an olive tree (Olea europaea) from the southern Peruvian arid coast was obtained for the first time. Genomic DNA of high quality was used to generate librarieswith Illumina Hiseq paired-end methods. The cp genome is 155,886 pb in length and contains a large single-copy region (LSC) of 86,610 pb and a small single copy region (SSC) of 17,790 pb separated by two inverted repeat (IR) regions (25,741bp). The cp genome of olive contains 124 genes that consists of 80 protein-coding genes, 36 tRNA, eight rRNA. Phylogenetic analysis showed this olive tree is sister to O. europaea subsp. maroccana (Oleeae tribe). This study presents the first overview of the chloroplast genome organization and phylogenetics of O. europaea, offering valuable insights for genetic and evolutionary research in the genus Olea.
plastome, NGS, coastal zone, phylogenomics
The olive Olea europaea (Oleaceae) is a subtropical tree distributed on most continents in tropical and temperate environments (Dupin et al., 2020). The genus Olea comprises about 40 old world species (Jensen et al., 2002) and the majority of Oleaceae species possess an economic importance as most olive fruits are used for oil extraction. Vegetable oil is important for its nutritional and healthy advantages compared to others (Rallo et al., 2000). Currently, ten olive tree chloroplast (cp) genomes were reported from Mediterranean region (Besnard et al., 2011), Italy, Spain, China (Niu et al., 2020), and Jordan (Haddad et al., 2021). However, to date, the olive tree chloroplast genome from Peru has not been reported. In this study, we sequenced, assembled, and annotated for the first time the complete chloroplast genome of a centennial the olive tree from Peru ( Figure 1), using next generation sequencing (NGS), providing valuable information for genetic and evolutionary studies in the genus Olea.
Plant materials and extraction of genomic DNA Fresh young leaves were collected from “El Algarrobal” district located in the Ilo province from Moquegua region (latitude 17.61294, longitude -71.27136). The specimen (MOQ001169) was deposited at the National University of Moquegua Herbarium (http://www.hmoqueguensis.unam.edu.pe/, Hibert Huaylla-Limachi hmoqueguensis@unam.edu.pe) under the voucher Nro. 001169. DNA was extracted using CTAB method (Doyle and Doyle 1990) with minor modifications for this specie. The quality and the quantity were evaluated on a 1% agarose gel and fluorescence using the Qubit™ 4 Fluorometer (Invitrogen, Waltham, MA, USA), respectively.
DNA sequence and genome assembly Library construction pair-end reads and sequencing were carried out using Illumina HiSeq 2500 platform and a PE 150 library, with the NexteraXT DNA Library Preparation Kit (Illumina, San Diego, CA, USA). We removed the adapters and verified the quality of reads employing Trim Galore (Martin, 2011) with arguments: −F embplant_pt −R 15 –reduce-reads-for-coverage inf. The cp genome was assembled with GetOrganelle v1.7.2 (Jin et al., 2020), using Olea europaea subsp. europaea (NC_015401) as reference. SPAdes v3.11.1 (Bankevich et al., 2012), bowtie2 v2.4.2 (Langmead and Salzberg, 2012), and BLAST+ v2.11 (Camacho et al., 2009) were also used in the pipeline with default settings.
The cp genome was annotated using GeSeq in CHLOROBOX web service (Tillich et al., 2017) Default settings were applied, and comparisons were made with all available plastid genomes of Oleae in the NCBI database, followed by manual curation
To understand the phylogenetic position of Olea europaea, a maximum likelihood (ML) tree of 19 genomes retrieved from GenBank was reconstructed. First, we employed MAFFT v.7.475 (Katoh and Standley, 2013) to align those genomes. Then, with a GTR + GAMMA model of evolution, we obtained the best-scoring ML tree, considering 1,000 bootstrap (BS) inferences with RAxML v8.2.11 (Stamatakis, 2014). We employed capirona (Calycophyllum spruceanum) cp genome as an outgroup (OK326865). The aligned data that was employed for the phylogenetic analysis consists of 187,111 bp (Supplemental Data 1, https://doi.org/10.5061/dryad.tmpg4f57q).
The complete chloroplast genome of olive was 155,886 bp in length with typical quadripartite structure that included a large single-copy (LSC) of 86,610 bp and a small single copy region (SSC) of 17,790 bp separated by two inverted repeat (IR) regions (25,741 bp). Following of annotation and subsequent modifications, we submitted the complete chloroplast (cp) genome sequence to the GenBank database. This submission is associated with the accession number: ON767107. The average depth of coverage is 17,673.05X (Supplementary Figure 1, https://doi.org/10.5281/zenodo.14061216). The chloroplast genome contains 124 genes that consisted of 80 protein-coding genes, 36 tRNA, 8 rRNA. Of the total genes reported 12 present one intron. PafI and clpP1 genes contained two introns. We further report that most genes are present as a single copy, except 14 genes that were duplicated in IR regions ( Figure 2). A total of 11 cis-splicing genes (rps16, atpF, rpoC1, pafl, clpP1, petB, petD, rpl16, rpl2, ndhB, ndhA) were identified; one trans-splicing gene (rps12) was also identified (Supplementary Figure 2, Supplementary Figure 3).
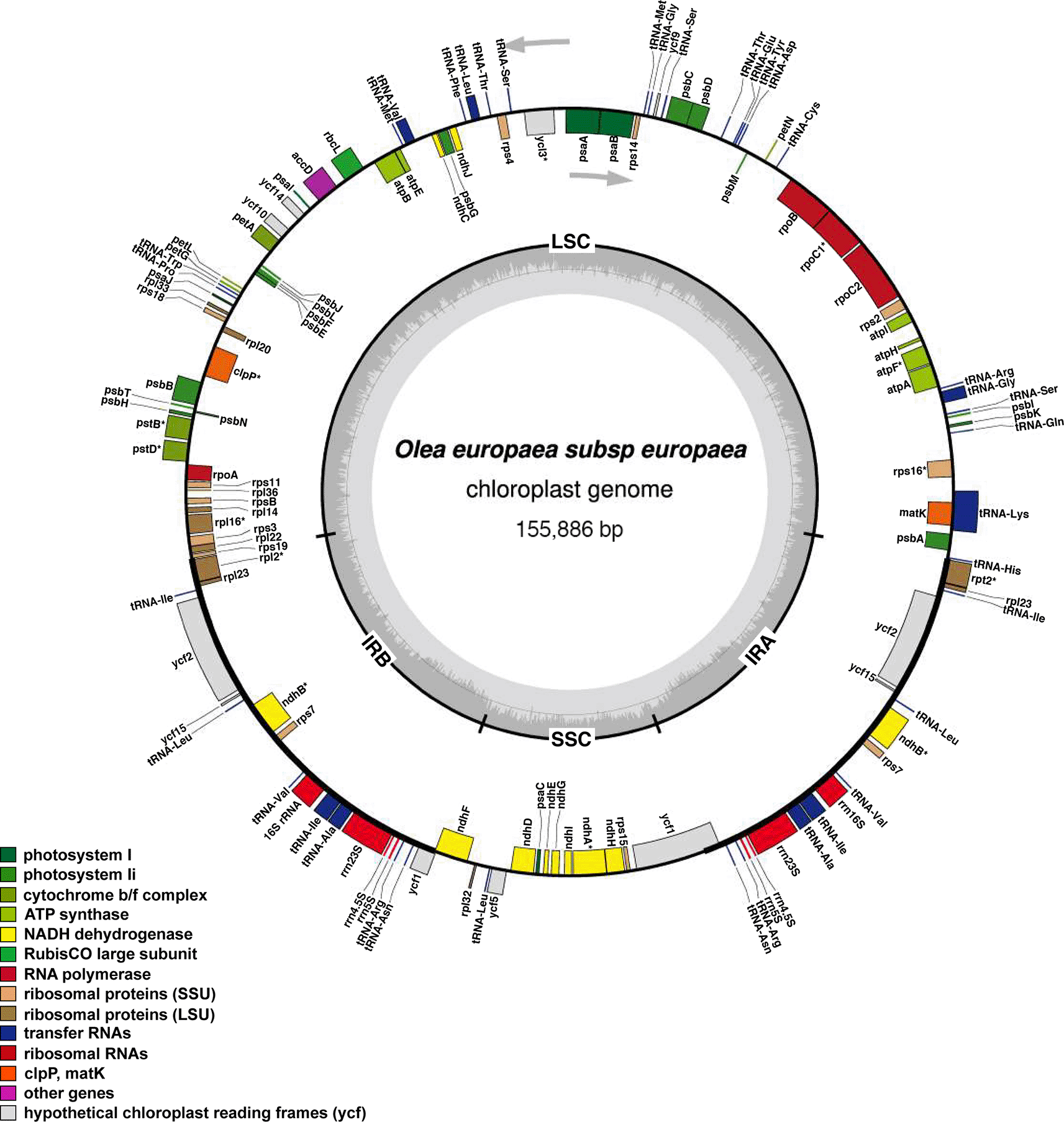
Genes belonging to different functional groups are color-coded. The genes functions are indicated colored in the bottom left corner. The dark grey inner circle indicates the presence of nodes in the LSC, SSC, IR regions).
This study examined 19 species of the Oleaceae family, with one outgroup, Calycophyllum spruceanum (OK326865), to understand their evolutionary relationships by analyzing complete chloroplast genome sequences. Maximum likelihood (ML) analysis revealed three distinct monophyletic groups within the tribes Jasminenae, Oleeae, and Forsythiae, with 17 of the nodes showing 100% bootstrap support. In addition, O. europaea is shown to be sister to O. europaea subsp. Maroccana, and sister to them is O. europaea subsp. cuspidata ( Figure 3). As expected, Olea europaea was categorized in the Oleeae tribe. These phylogenetic trees are consistent with the established classification of the Oleaceae family.
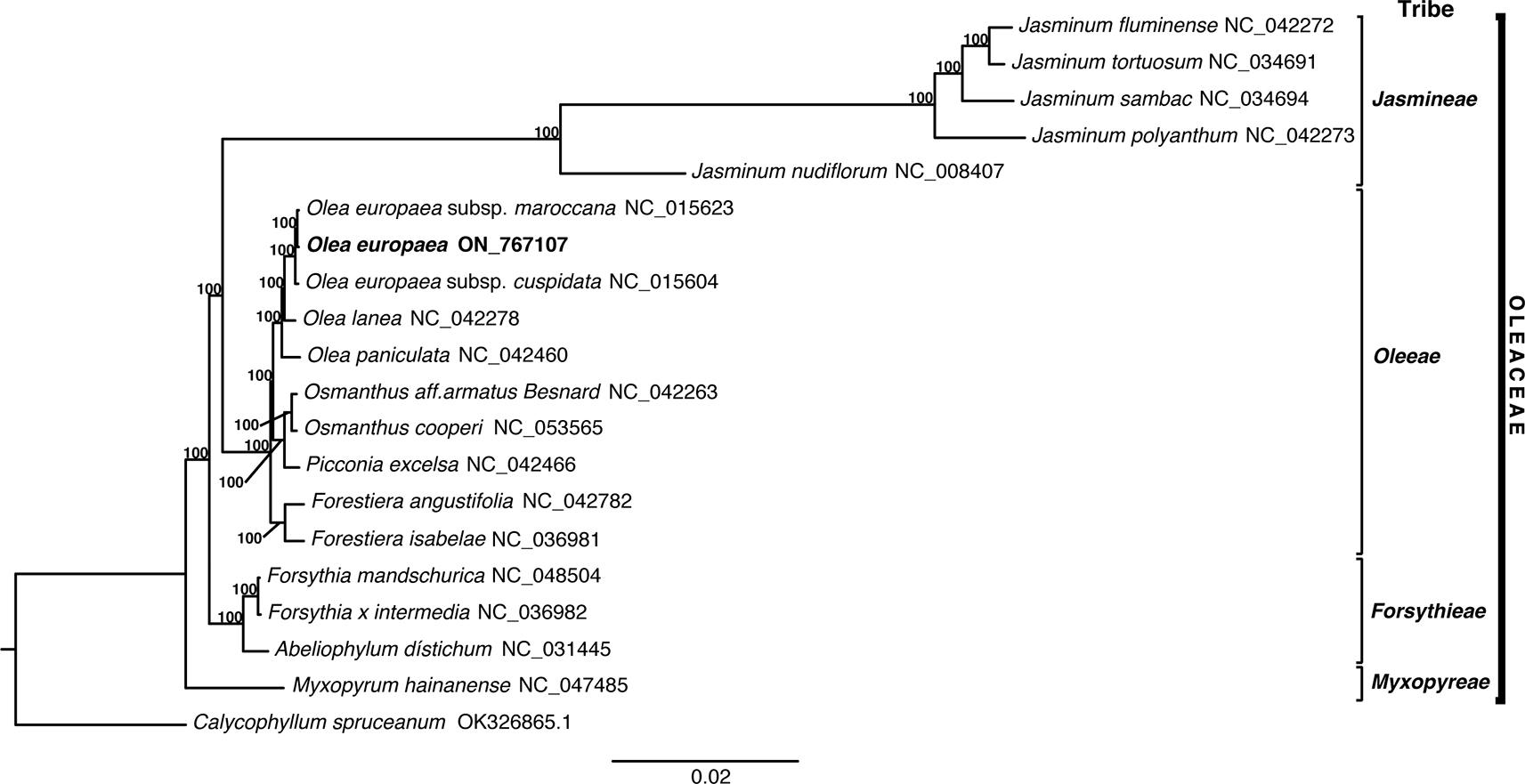
In this study, we assembled for the first time the chloroplast genome sequence of the centennial olive tree (Olea europaea) from the southern Peruvian coast. The results differ from other chloroplast genomes of Olea europaea, such as O. europaea subsp. europaea (NC_015401), which presents 85 protein-coding genes, 37 tRNA, and 8 rRNA (Haddad et al., 2021). Niu et al. (2020) reported that two varieties of O. europaea, subsp. europaea var. sylvestris and subsp. cuspidata, presented 133 genes, including 87 protein-coding genes, 37 tRNA, and 9 rRNA. Likewise, they found that the chloroplast genome length for both species were 155,886 bp (Niu et al., 2020). The phylogenetic results showed O. europaea is sister to O. europaea subsp. maroccana and O. europaea subsp. cuspidata in independent sister clade to other species of genus Olea the chloroplast genome sequence and annotation were submitted to NCBI with accession number ON767107.1.
Peruvian olive chloroplast genome will stimulate additional work to develop molecular markers for their application in a genomic selection program of superior individuals in early stages. In addition, this study may be a basis to know the location of genes of interest that may be used in gene editing. This work promotes knowing more about the genetics of the Peruvian olive tree, promoting its modern genetic improvement and conservation.
C.L.S. analyzed the data, was also involved in drafting the article. L.C.-L., C.I.A. and R.E. analyzed the data. E.F.-H., F.Z.-V., J.C.G.-A., C.A.-G., D.L.G.-R. were involved in the conception and design of the work. L.C.-L. and F.Z.-V. were involved in sample collection. F.Z.-V., J.C.G.-A., P.I. and C.I.A. were involved in data interpretation.
F.Z.-V., J.C.G.-A., P.I. and C.I.A. were involved in funding acquisition. C.I.A was involved in drafting the article. All authors read and approved the manuscript.
Olive sample collected and employed in this work do not involve protection determined by the Republic of Peru. Consequently, this study was exempted from ethical approval.
NCBI: Complete chloroplast genome. Accession number ON767107; https://www.ncbi.nlm.nih.gov/nuccore/ON767107.
Dryad: Supplementary material.
The complete chloroplast genome of a centennial olive tree (Olea europaea, Oleaceae) from the southern Peruvian coast. https://doi.org/10.5061/dryad.tmpg4f57q (Saldaña et al., 2024a).
This link contains the following extended data:
Zenodo: Supplementary material. https://doi.org/10.5281/zenodo.14061216 (Saldaña et al., 2024b).
This link contains the following extended data:
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication) (http://creativecommons.org/publicdomain/zero/1.0/).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Are the rationale for sequencing the genome and the species significance clearly described?
Partly
Are the protocols appropriate and is the work technically sound?
Partly
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genomics, Proteomics and bacteriocins, smart probiotics
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Genome assembly using long-read sequencin
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 04 Dec 24 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)