Keywords
Anopheles gibbinsi, understudied anopheline, genome skimming, Zambia
Mosquitoes belonging to the genus Anopheles are the only vectors of human malaria. Anopheles gibbinsi has been linked to malaria transmission in Kenya, with recent collections in Zambia reporting the mosquito species exhibiting zoophilic and exophilic behavioral patterns with occasional contact with humans. Given the paucity of genetic data, and challenges to identification and molecular taxonomy of the mosquitoes belonging to the Anopheles genus; we report the first complete mitochondrial genome of An. gibbinsi using a genome skimming approach. An Illumina Novaseq 6000 platform was used for sequencing, the length of the mitochondrial genome was 15401 bp, with 78.5% AT content comprised of 37 genes. Phylogenetic analysis by maximum likelihood using concatenation of the 13 protein coding genes demonstrated that An. marshallii was the closest relative based on existing sequence data. This study demonstrates that the skimming approach is an inexpensive and efficient approach for mosquito species identification and concurrent taxonomic rectification, which may be a useful alternative for generating reference sequence data for evolutionary studies among the Culicidae.
Anopheles gibbinsi, understudied anopheline, genome skimming, Zambia
A few species in the genus Anopheles are well established primary vectors of human malaria in sub-Saharan Africa, including Anopheles gambiae and An. funestus. However, studies have also implicated understudied anopheline mosquito species in driving transmission in regions where primary vectors are close to elimination.1–3 Anopheles gibbinsi, until recently recognized as An. species 6, has tested positive for Plasmodium falciparum sporozoites in studies from Kenya.4,5 Furthermore, An. gibbinsi was previously reported from central, eastern and northern Africa and has now been shown to have a geographic range extending into southern Africa. Recent first-time captures for this species in Zambia reported the species largely exhibiting zoophilic and exophilic behavioral patterns; however, a blood meal PCR assay also detected a few specimens positive for human host DNA.5 Similar in morphology to other well-established malaria vectors and with a dearth of genetic data available, there is a need for the continued monitoring of An. gibbinsi as a potential vector in malaria transmission. It is also important that tools for accurate mosquito identification be developed, due to limitations with the commonly targeted cytochrome oxidase I gene (COI) and the internal transcribed spacer 2 (ITS2) in resolving members of morphologically cryptic species complexes.6
The expansion of sequencing strategies has employed the use of mitochondrial genomes (mitogenomes) primarily for species identification and solving discrepancies in the taxonomic classification of metazoan organisms. Mitogenomes have also proved useful in evaluating population structure, chromosomal rearrangements, species introgression, and evolutionary histories.7–9 The mitochondrial DNA (mtDNA) is a circular double stranded molecule that encodes 37 genes made up of 13 protein coding genes (PCGs), 22 transfer RNA (tRNA) genes), 2 ribosomal RNA (rRNA) genes and an adenine and thymine (A-T) rich terminal, an essentially non-coding area termed the control region (CR) associated with the replication and transcription of the genome. Maternal inheritance, high copy number, lack of recombination, and absence of introns are characteristics which allow mtDNA to be well suited for accurate molecular identification and rectifying taxonomic classification among species.6,10 Here for the first time, we describe a genome skimming approach for recovery of, and characterization of the mitochondrial genome of An. gibbinsi and its phylogenetic relationship to other established anopheline vectors of human malaria.
The An. gibbinsi specimens (n = 3) sequenced were collected in Nchelenge, Zambia using a CDC light trap that was placed near an animal. The specimens were stored on silica gel until DNA extraction. Single mosquito specimens were pre-treated11 as described by Chen et al. 2021, followed by the extraction protocol as per manufacturer’s instructions (Qiagen DNeasy Blood and Tissue Kit, Hilden, Germany). A whole mosquito specimen was placed in a 1.5 mL Eppendorf tube and homogenized in a cocktail containing 98 μL of PK buffer (Applied Biosystems, Waltham, Massachusetts, U.S.A.) and 2 μL of Proteinase K (100 mg/mL), this was followed by an incubation step for 3 hours at 56oC.12 After incubation, 100 μL of isopropanol and 100 μL of Buffer AL (Qiagen DNeasy Blood and Tissue Kit, Hilden, Germany) was added to the lysate and left to incubate at room temperature for 10 minutes. The mixture was pipetted into a DNeasy mini spin column placed in a 2 mL collection tube; extraction protocol was followed5and stored at -20oC prior to sequencing. DNA was shipped to SeqCenter (Pittsburg, U.S.A) for library construction and sequencing. Libraries were sequenced from both ends (150 bp) on Illumina Novaseq 6000 to a depth of 13.3 million reads.
The mitochondrial genome contigs were assembled similar to that of the An. squamosus13 mitogenome using NOVOPlasty (RRID:SCR_017335) version 4.3.1.12 Using the invertebrate genetic code under default settings, automatic annotations were conducted using the MITOS website.14 Adjustments for start and stop codon positions were performed manually in Geneious Prime (RRID:SCR_010519) version 2023.2.1 (Biomatters, Auckland, Australia) to match reference anopheline mitogenomes deposited in NCBI’s GenBank database. The mitochondrial genome sequences and their corresponding annotations were submitted to the GenBank database. A representative mitogenome map is provided in Figure 1.
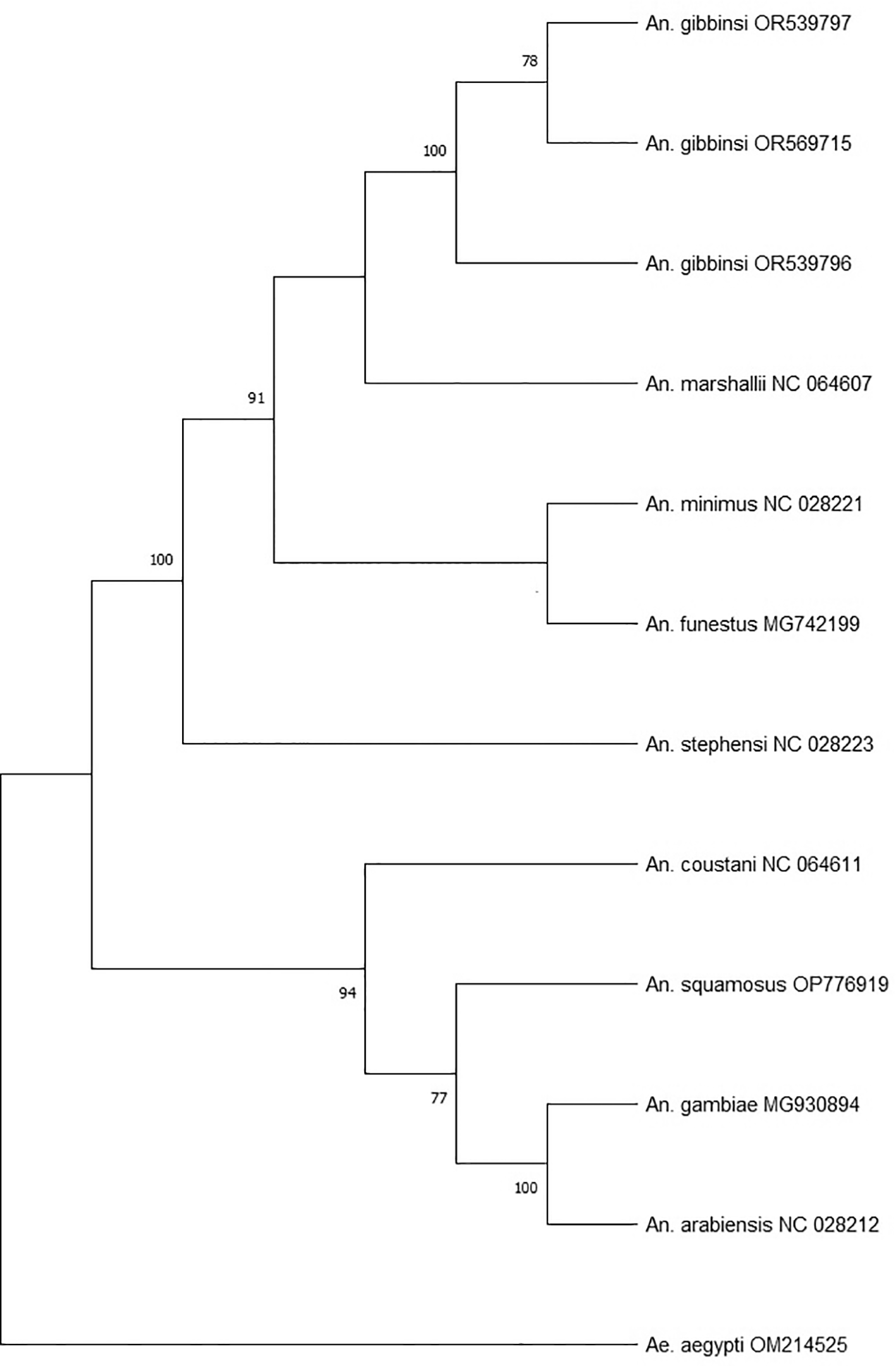
Phylogenetic analysis was performed using the concatenated 13 PCGs of the three An. gibbinsi specimens, eight Anopheles species and one Aedes species as an outgroup. The General Time Reversible (GTR + G + 1) model was identified as the best fit for building the maximum likelihood phylogenetic tree in MEGA (RRID_SCR_023017) version 1115 using 1000 bootstrap replicates.
The sequencing from the 3 An. gibbinsi samples yielded an average of 29,674,206 million reads and of these, approximately 119,364 reads were used to assemble each mitochondrial genome. The contents of the 3 An. gibbinsi mitogenomes (GenBank accession numbers OR_539796, OR_539797, OR_569715) included 2 ribosomal RNAs, 22 transfer RNAs and 13 protein coding genes. The representative mitochondrial genome (OR_539797) length was 15,401 bp with an A + T percentage of 78.5% which is comparable to other anopheline mitogenomes deposited in the GenBank database. The cytochrome c oxidase I (COI) fragment spanning 8598–10,133 bp was 94.5% similar to a COI sequence for An. marshallii (GenBank YP_010419919).
Phylogenetic analysis (Figure 2) using the concatenated PCGs revealed An. marshallii (NC_064607) as the closest sequenced relative to An. gibbinsi, forming a single but weakly supported clade apart from the well-recognized vectors of malaria. Both species belong to the An. marshallii complex.
Genome skimming has demonstrated to be a cost-effective approach for generating reference sequence data which can be used for mosquito identification and resolving phylogenies. With the continued monitoring of An. gibbinsi as a potential vector for malaria transmission, this study provides a key genomic resource for understanding the phylogenetic relationship of this mosquito species within its complex and with primary vectors of human malaria transmission.
GenBank: Anopheles gibbinsi mitochondrion, complete genome. Accession numbers OR539796, OR539797, OR569715. 16
Bio Project. Complete mitogenome sequence of Anopheles gibbinsi from Nchelenge, Zambia, Accession number PRJNA1072262. 17
SRA. Illumina seq of Anopheles gibbinsi. Accession numbers SRR27842795, SRR27842796, SRR27842794. 18
BioSample: Anopheles gibbinsi isolates ANGB, AGB1, GN. SAMN39739077, SAMN39739078, SAMN39739079. 19
We would like to thank the Zambian field teams and the Tropical Diseases Research Centre for assisting with sample collection. Many thanks to Dr. Yoosook Lee, University of Florida for guidance with bioinformatic analysis.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: comparative genomics
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Partly
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Population genetics of Anopheles mosquitoes.
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Partly
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: My research area is population genomics and molecular phylogenetics. I have worked on Aedes species mitochondrial phylogeny and related flaviviruses.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 30 May 24 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)