Keywords
Obesity, Overweight, LEP rs2167270, LEPR rs1137100, genetic variants, Jordan
This article is included in the Agriculture, Food and Nutrition gateway.
Previous Genome Wide Association Studies (GWAS) of obesity susceptibility genes in different populations have confirmed the association of some variants with obesity, body mass index (BMI), and some related metabolic traits. To our knowledge, the current study is the first to investigate the genetic basis of obesity in the Jordanian population. The aim of our study is to investigate the occurrence and frequency of obesity-related genes in Jordanian individuals and any possible relationship between SNP genetic markers and phenotypic characteristics of studied individuals.
A total of 150 Jordanian unrelated adults, including 83 obese, 26 overweight, and 41 normal-weight subjects, were genotyped using the high resolution melt analysis (HRMA) and tested for the association of single nucleotide polymorphisms (SNPs), rs2167270 in LEP gene and rs1137100 in LEPR gene, with obesity risk, obesity/overweight risk, BMI, weight, height, total cholesterol, HDL-cholesterol, LDL-cholesterol, triglycerides, and blood level glucose.
A significant association between obesity risk and the rs2167270 mutation in LEP gene was observed under a dominant model (odds ratio (OR) = 2.5147, 95% CI =1.0629 to 5.9494, p = 0.0358). A marginal (not significant) association between BMI and the rs2167270 mutation in LEP gene (p = 0.075), was also detected. Moreover, an association between rs2167270 mutation in LEP gene and glucose blood level was observed (p = 0.038). Total cholesterol and LDL-cholesterol also presented a weak association with the rs1137100 mutation in LEPR gene, with p = 0.071 and p = 0.078, respectively. Additionally, no significant association between LDL-cholesterol and the rs2167270 mutation in LEP gene was observed (p = 0.091). By contrast, there was no association between weight or height and the SNPs mutation evaluated in this study (p > 0.05).
These results suggest that some SNPs in some obesity-related genes may contribute to obesity risk and its related anthropometric and metabolic traits in Jordanian population. To confirm these results, further studies on a larger Jordanian cohort should be carried out.
Obesity, Overweight, LEP rs2167270, LEPR rs1137100, genetic variants, Jordan
Obesity is a multi-factorial disease resulting mainly from environmental and genetic factors, are defined as ‘abnormal or excessive fat accumulation that presents a risk to health’. A body mass index (BMI) over 25 is considered overweight, whereas over 30 is obese.1 According to the latest update of the Human Obesity Gene Map there are 253 quantitative trait loci related with obesity and obesity-related phenotypes that were identified from 61 genome-wide scans. Additionally, 127 candidate genes were associated with obesity phenotypes.2 Moreover, it has been reported that in genome wide association studies (GWAS) the authors analyze the interaction between genetic predisposition and the intake of sugar-sweetened beverages in relation to body-mass index, based on these 32 loci that were previously reported.3,4 Six new loci associated with BMI highlight a neuronal influence on body weight regulation,4 and association analyses of 249,796 individuals reveal 18 new loci associated with BMI.4,5 Association analyses of 249,796 individuals reveal 18 new loci associated with BMI.5 Leptin, a 16-kDa hormone of 167 amino acids, is encoded by LEP gene on chromosome 7,6 and is secreted mainly by the adipose tissue,7 is a member of the leptin-melanocortin pathway. Leptin plays a key role in energy homeostasis and in the control of satiety. Leptin exerts this action by binding to its receptor (LEPR) in the hypothalamus in a region responsible for energy homeostasis and appetite.8 Based on this mode of action, variations in the LEP or LEPR genes may have a role in the development of obesity or its markers.
Studies evaluating the occurrence of single nucleotide polymorphisms (SNPs) in particular genes can provide insights into how changes in the genome may affect the risk of developing a disease such as obesity. SNPs are variations in a single nucleotide base in DNA that can occur between individuals, and some of these variations have been linked to an increased or decreased risk of developing certain diseases.9
For example, certain SNPs in genes that regulate appetite and energy expenditure have been found to be associated with an increased risk of obesity. These genes may play a role in how the body regulates food intake and energy balance, and variations in these genes may alter these processes, leading to an increased risk of developing obesity.10 By identifying specific SNPs associated with obesity risk, researchers can gain a better understanding of the genetic factors involved in the development of this disease. This knowledge can also be used to develop personalized approaches to preventing and treating obesity based on an individual’s genetic profile.5 However, it’s important to note that genetic factors alone cannot fully explain the development of obesity, and other factors such as lifestyle, environment, and epigenetic modifications also play a role.11
To our knowledge, this study is the first to investigate the putative genetic basis of obesity in the Jordanian population. This study aims to investigate if there is a relationship between mutation in the genome and the obesity risk in Jordanians through the analysis of two variants in leptin and leptin receptor genes namely, rs2167270 and rs1137100, respectively, and to investigate any possible relationship between these two variants and phenotypic characteristics of studied individuals.
The study was approved by the Institutional Committee of the faculty of graduate studies at Al-Balqa Applied University on March 1, 2019 (Proposal Reference Number: session 6/2019). Human procedures were followed in accordance with the Helsinki Declaration of 1975 as revised in 2013. Written informed consent was obtained from all patients, indicating their agreement to take part in the study.
A total of 150 Jordanians with no other diagnosed disease between the ages of 19-55 years participated in this study during the period of March and July 2019. The study was performed on volunteers from Al-Balqa Governorates and executed at Al-Balqa Applied University. Data on dietary habits, medical implication, physical activity, and family history for each participant were obtained by completing an interview questionnaire (face-to-face interactions).
Inclusion criteria include that the person belonged to the University, either students, academic teachers or members of the administration body. The exclusion criteria encompassed individuals with heart diseases, kidney issues, and metabolic syndrome. It also involved those who underwent bariatric surgeries or any stomach-related intervention for weight reduction, as well as individuals whose overweight or obesity result from hormonal disorders (i.e. hypothyroidism, Cushing’s disease, and hypogonadism) or the use of cortisone or other medications.
Height was measured with a portable Harpenden stadiometer® to the nearest millimeter (0.1 cm). Participants were barefoot, dressed in minimal clothing, standing with heels together, arms out to the side, legs straight, shoulders relaxed and the head in the Frankfurt plane. Body weight was measured using an Inbody™ 770 scale, to the nearest 0.1 kg with the subjects barefoot and wearing light clothes. BMI was calculated using the formula Weight/Height (kg/m2) and study participants were categorized normal (BMI 18.5–24.9) as overweight (BMI ≥ 25) and obese (BMI ≥ 30 according to the World Health Organization (WHO) standards for adults.12
Peripheral venous blood samples after 10-12 hours fasting were collected in EDTA and plain tubes for SNPs genotyping and serological tests, respectively. Blood samples collected in plain tubes were centrifuged at 2,500 rpm for 15 minutes. The resulting serum was transferred to Eppendorf tubes and stored at -20°C until they were used for the serological analyses within a period not exceeding three months.
Serum lipid levels, and fasting serum glucose (FSG) were analyzed with test kits (Biolabo) and expressed as mg/dl. Absorbance was measured using a Biovawe II spectrophotometer. FBG was determined by the glucose oxidase method and total cholesterol (TC) and triglyceride (TG) concentrations were determined using standard enzymatic methods. High density lipoprotein-cholesterol (HDL-C) was measured by precipitation method. Finally, low density lipoprotein-cholesterol (LDL-C) was calculated using Friedewald formula.
Genomic DNA was extracted from white blood cells contained in 1.0 ml peripheral blood using Wizard® Genomic DNA Purification Kit (Promega) according to manufacturer’s instructions. DNA quality and concentration were assessed using a gel electrophoresis and a Biovawe II spectrophotometer. DNA solutions were prepared with a final concentration of 50 ng/ul and stored at -20°C until use.
Primers were designed to flank the selected SNP using Primer3 (RRID:SCR_003139) software applying the following criteria: product size: 40-100 bp, length: 19-23 bp, GC content: 45-60%, GC clamp: up to 3, annealing temperature: 55-60°C and an annealing temperature difference between each two pairs of primers of 1°C. The quality of the chosen primers, primer-dimer or cross dimer was assessed using DNAMAN software DNAMAN 10.0 – Integrated System for Sequence Analysis13 and Primer Premier 6.25 – PCR Primer Design Software.14 For the optimization of primers annealing temperature, PCR reactions for each pair of primers were performed at three different annealing temperatures above and below the calculated Tm (e.g., 50, 55 and 60°C, when the calculated Tm was 55°C). The optimal annealing temperature was the one that gave the lowest number of quantification cycles (Cq), with no nonspecific amplification or primer dimers formation. Then, the PCR product was evaluated using an 0.8% agarose gel electrophoresis and the PCR product with one single, bright band of the expected size on was chosen to be the optimized to PCR amplification. The selected primers used in this study are listed in Table 1. PCR amplification was carried out using a master mix from Kappa-bio Systems, prepared to fit the requirement of the High-Resolution Melt Analysis (HRMA) technology, in which a special dsDNA dye (EvaGreen) was included. The selected primers, related to each SNP, were used to amplify the region containing the respective SNP. DNA amplification was carried out using CFX96 real-time PCR applying the following cycling parameters: initial denaturation at 95.0°C for 4:00 min, denaturation at 95.0°C for 0:10 min, primers annealing step for 0:30 min (at different temperature according to the results of the primer annealing temperature optimization for each SNP as shown in Table 1). The extension step was carried out at 72.0°C for 0:30 min. A total of 37 cycles were used for both SNP amplification. The amplification step was followed by the melting step by raising the temperature from 65.0°C to 95.0°C (increment 0.5°C, 0:05) after a denaturation step at 95°C for 1:00 minute. The resulting melt curves were analyzed using Precision Melt Analysis™ software version 1.3.15 The SNPs for the selected genes were genotyped using the HRMA technique. Data were generated using CFX96 real-time PCR detection system and analyzed using Precision Melt Analysis™ software.
Statistical analysis was performed using IBM SPSS Statistics (RRID:SCR_016479), version 23 (SPSS Inc., Chicago, IL, USA). The association of each SNP with the risk of obesity was assessed by calculating Person’s chi-square test and odds ratio (OR). The analysis was performed under the dominant, recessive, and co-dominant models. The associations of BMI and metabolic traits with genotypes were evaluated using the analysis of variance test (ANOVA) using t-test. P-values < 0.05 were considered statistically significant.
The biochemical and clinical characteristics of the participants in this study are summarized in Table 2. A total of 83 participants were identified as obese, 26 as overweight, and 41 as normal weight. There were significant differences in the BMI (p < 0.0005), weight (p < 0.0005), triglycerides (p = 0.005) between obese, overweight, and normal weight groups. Among the participants with obesity, 46 were within class I obesity, 20 within class II, and 17 within class III. BMI means were 32.3 ± 3.3, 36.6 ± 4.7, and 44.6 ± 7.4 for class I, class II and class III respectively.
The genotyping success rate for LEP rs2167270 for the 150 participants was 87% (131 participants were successfully genotyped), of which 41 (31.3%) were homozygous for the wild-type (GG) genotype, 59 (45.0%) were heterozygous (GA), and 31 (23.7%) were homozygous for the variant (AA) genotype (Figure 1). The genotypes of this SNP were in Hardy-Weinberg equilibrium (χ2 = 1.16, p > 0.05). The frequency of the rs2167270 (A) allele (minor allele frequency) for all participants was (46%). There was a significant difference (p ≤ 0.05) in the risk allele (A) frequency within normal, obese, and obese/overweight participants. The frequency for normal weight was 41%, for obese was 53%, and for the obese/overweight group was 48%.16
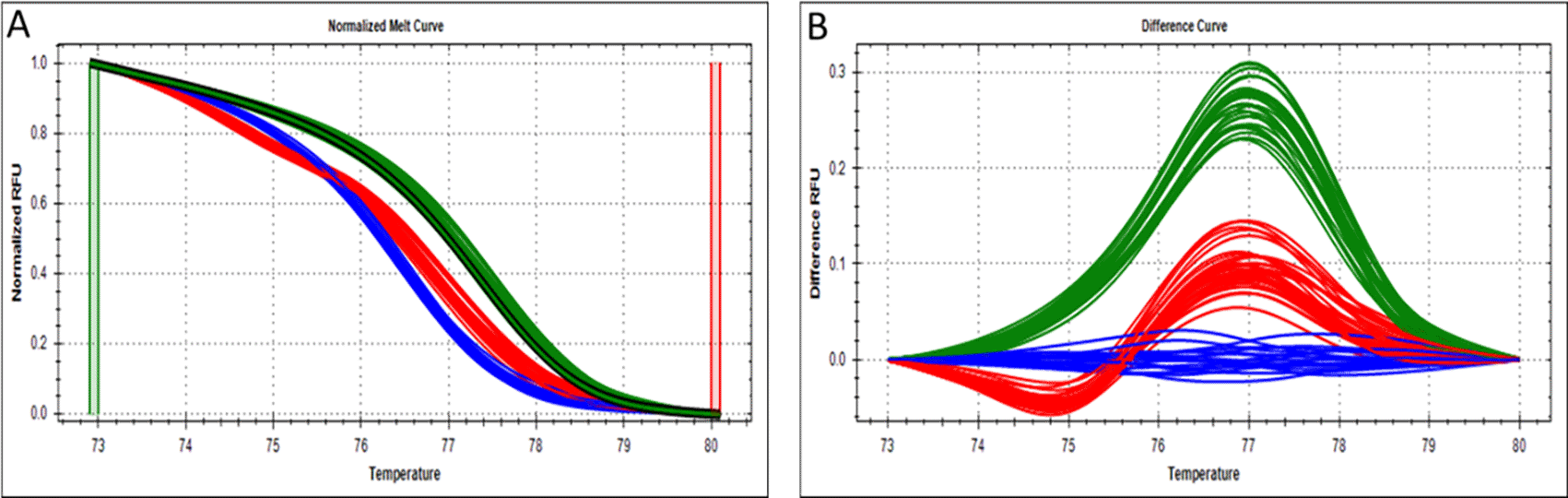
Data were generated using CFX96 real-time PCR detection system and analyzed using Precision melt analysis software. (A) Normalized melt curves. (B) Difference curves. GG genotype (wild type) is represented by the green curves cluster. AA genotype (mutant) is represented by the blue curves cluster. GAgenotype (heterozygote) is represented by the red curves cluster. (Each curve represents one single sample).
The mean values of BMI for GG, GA, and AA genotypes were 28.4, 31.6, and 31.2, respectively. ANOVA test was applied to evaluate the association between rs2167270 genotype and the anthropometric and metabolic characteristics. No association was observed between this SNP and the BMI, although a certain trend was detected (p = 0.075; Table 3). No other associations were observed with the BMI within the different participants groups considered but a certain trend toward significance within the female subgroup was determined (p = 0.09; Table 4). A significant association between the LEP rs2167270 with the fasting blood glucose levels (p = 0.038) was found. However, no associations were observed with the other metabolic traits evaluated in this study (Table 3). However, an association was found between the LEP rs2167270 and obesity under the dominant model (AA + GA vs. GG; OR = 2.52, 95% CI = 1.063-5.949, p = 0.036; Table 5). The Chi-square test was performed and a trend toward significance was found between the LEP rs2167270 and the risk of obesity under the additive model (χ2 (2, N = 109) = 4.895, p = 0.086; Table 5). Using the same model, no association was observed between this variant and the obesity/overweight risk, for all participants (χ2 (2, N = 131) = 4.09, p = 0.130; Table 6) nor for male or female subgroups (Table 7). The association between the LEP rs2167270 and the risk of obesity/overweight under the dominant and the recessive mutations (AA vs. GA + GG) was also tested (Table 6). No association was detected within all participants under these models, although the dominant model showed a certain trend toward significance (OR = 2.019, 95% CI = 0.901-4.524, p = 0.088; Table 6).
| LEP rs2167270 genotype | |||||
|---|---|---|---|---|---|
| GG | GA | AA | All | p-value | |
| Body mass index (kg/cm) | 28.4 ± 1.1 (41) | 31.6 ± 0.9 (59) | 31.2 ± 1.4 (31) | 30.5 ± 0.6 (131) | 0.075 |
| Body mass index (kg/cm) males only | 29.4 ± 1.4 (27) | 31.5 ± 1.2 (33) | 32.3 ± 1.7 (19) | 31.0 ± 0.8 (79) | 0.356 |
| Body mass index (kg/cm) females only | 26.5 ± 1.4 (14) | 31.7 ± 1.4 (26) | 29.5 ± 2.4 (12) | 29.8 ± 1.0 (52) | 0.090 |
| Height (cm) | 169.2 ± 2.0 (29) | 169.0 ± 1.3 (50) | 167.7 ± 1.7 (26) | 168.7 ± 0.9 (105) | 0.806 |
| Weight ( kg) | 88.4 ± 4.8 (29) | 91.6 ± 3.1 (50) | 91.1 ± 5.1 (26) | 90.6 ± 2.3 (105) | 0.843 |
| Fasting serum glucose (mg/dl) | 80.6 ± 1.9 (19) | 87.7 ± 1.7 (29) | 81.6 ± 5.1 (16) | 84.1 ± 1.3 (64) | 0.038* |
| Total cholesterol (mg/dl) | 215.1 ± 10.1 (24) | 194.6 ± 9.2 (38) | 191.1 ± 12.8 (20) | 199.7 ± 6.1 (82) | 0.265 |
| Triglycerides (mg/dl) | 112.0 ± 11.0 ( 24) | 124.5 ± 13.2 ( 38) | 140.7 ± 21.7 (20) | 124.8 ± 8.7 (82) | 0.488 |
| HDL (mg/dl) | 41.4 ± 3.1 (21) | 39.2 ± 1.9 (38) | 38.7 ± 2.7 (19) | 39.7 ±1.3 (78) | 0.747 |
| LDL (mg/dl) | 158.6 ± 9.7 (21) | 130.5 ± 9.3 (38) | 123.9 ± 13.3 (19) | 136.5 ± 6.3 (78) | 0.091 |
| Obese only | Normal-weight | OR (95% CI) | p-value | |
|---|---|---|---|---|
| Dominant model | ||||
| N | 74 | 35 | 2.5147 (1.0629 to 5.9494) | 0.0358* |
| GG | 17 (23%) | 15 (42.9%) | ||
| AA+GA | 57 (77.0%) | 20 (57.1%) | ||
| Recessive model | ||||
| N | 74 | 35 | 1.1447 (0.4603 to 2.8467) | 0.7713 |
| AA | 21 (28.4%) | 9 (25.7%) | ||
| GG + GA | 53 | 26 | ||
| Additive model | ||||
| Pearson Chi-Square value | p-value | |||
| N | 74 | 35 | 4.895 | 0.086 |
| GG | 17 (23%) | 15 (42.9%) | ||
| GA | 36 (48.6% | 11 (31.4%) | ||
| AA | 21 (28.4%) | 9 (25.7%) | ||
The genotyping success rate for LEPR rs1137100 was 94.7% (142 participants were successfully genotyped) of which 116 (81.7%) were homozygous for the wild-type (AA) genotype, 19 (13.4%) were heterozygous (GA), and 7 (4.9%) were homozygous for the variant (GG) genotype (Figure 2). The genotypes of this SNP were in Hardy-Weinberg equilibrium (χ2 = 17.25, p > 0.05). The minor frequency of the rs1137100 G allele for all (142) participants was 12%. There was a slight (not significant) difference in the risk allele (G) frequency within normal (12%), obese (13%), and obese/overweight participants (11%).

Data were generated using CFX96 real-time PCR detection system and analyzed using Precision melt analysis software. (A) Difference curves. (B) Normalized melt curves. AA genotype (wild type) is represented by the red curves cluster. GG genotype (mutant) is represented by the blue curves cluster. GA genotype (heterozygote) is represented by the green curves cluster. (Each curve represents one single sample).
The mean values of BMI for AA, GA, and GG genotypes were 31.0, 29.5, and 28.7, respectively. ANOVA test, which was applied to find the association between the LEPR rs1137100 genotype and the anthropometric and metabolic traits, showed no association of this SNP with BMI (p = 0.55; Table 8). However, marginal associations with total cholesterol (p = 0.071) and LDL cholesterol (p = 0.078) were observed (Table 8). Also, no associations were observed with BMI within participants groups (Table 9). The chi-square test was performed, and no relationship was found between the risk of obesity and the LEPR rs1137100 genotype using the additive model (X2 (2, N = 142) 0.26, p = 0.987). The association of LEPR rs1137100 with the risk of obesity using the dominant (AA vs. GA + GG), and recessive (AA + GA vs. GG) models were also tested (Table 9). No associations were detected within all participants groups using these models (Table 10), or within males or female separately (Table 11).
Since the discovery of leptin,6 many association studies have been carried out to identify the variations in the LEP gene that may be associated with obesity and its related anthropometric and metabolic characteristics.17,18 One of these variations was the rs2167270 (A19G), located in the first exon of the 5′ Untranslated Region.17 So far, few studies specifically concerning the association between the LEP rs2167270 genotype and obesity and its related anthropometric and metabolic characteristics are available.18–23 Table 2 shows a sample of these studies.
The genotype results for LEP rs2167270 obtained in this study are similar to the frequencies of the three LEP rs2167270 genotypes presented in several different populations studied in the International HapMap project.16
So far, most studies focused on the association of rs2167270 with serum leptin level, as this SNP might be involved in obesity by affecting leptin concentration.24 A systematic review and meta-analysis study25 addressing the associations of some adipokine genes variants with obesity susceptibility, revealed 10 studies that concerned with LEP variants. Four of these studies concerned the rs2167270 variant in particular.18–21 The overall results of these four studies, which included 553 obese and 365 controls, reported a non-significant association with obesity under dominant (OR = 1.14, 95% CI = 0.75–1.74 p = 0.53) and recessive models (OR = 0.89, 95% CI 0.67–1.20 p = 0.45). In the present study, the percentage of normal-weight participants who carried the GG genotype (42.9%) were larger than the percentage of obese/overweight participants (27.1%) who carried the same genotype, and the percentage of the normal-weight participants whose genotype is CC or CT (57.1%). It was also slightly smaller than the percentage of the obese/overweight participants (72.9%) whose carried these same genotypes. However, using the odds ratio under the dominant model, this difference did not reach the significant level of association with the risk of overweight/obesity, which is consistent with a Brazilian study22 and the four other studies included in the meta-analysis mentioned above, but it showed a certain trend toward significance (OR =2.019, 95% CI = 0.901-4.524, p = 0.0878), which was further verified to be restricted to the risk of obesity alone (OR =2.5147 95% CI =1.0629 to 5.9494, p = 0.0358). This result is consistent with an Indian study23 under dominant model (OR = 1.699, 95% CI =1.043–2.766, p = 0.03314). Association of LEP rs2167270 with obesity-related traits, i.e., BMI, has been the subject of many studies (Table 2) with contradictory results. While some studies confirmed the existence an association regarding to BMI,23,26 other studies revealed opposite results.18,20
The mean BMI of the participants not carrying any of the LEP rs2167270 risk alleles (GG) was 28.4, which is lower than the mean BMI of those carrying one risk allele (GA, 31.6), and carrying two risk alleles (AA, 31.2). This difference did not show a significant association but only a significant margin (p = 0.075), which may be due to the small sample size analyzed. However, these results are in line with those reported in other studies.18,20 Additionally, we observed that this small margin was maintained in the women’s group (p = 0.090) but was not observed when the men’s group was analyzed separately (p = 0.356), which suggests a Sex effect in this association between the LEP rs2167270 and the BMI. To validate these findings, studies involving a larger Jordanian cohort of each Sex should be conducted. To our knowledge, there are no studies reporting an association of LEP rs2167270 with serum lipid and glucose levels in adults, but a study including 136 Japanese children found no significant association of this SNP with serum lipids levels.27
The LEPR rs1137100 is a functional SNP in which the adenine (A) nucleotide is replaced by a guanine (G) nucleotide in exon two of the LEPR gene. As a result, lysine amino acid (K), encoded by the codon (AAG), is replaced by arginine (R), which is encoded by the codon (AGG). Rung et al., represents the distribution of the three LEPR rs1137100 genotypes in different populations studied in the International HapMap project.28 In general, it can be noted that the genotyping results for LEPR rs1137100 obtained in this study are similar to the genotyping frequencies in all populations, except for Japanese Tokyo (JPT), Han Chinese (CHB and HCB), and Chinese in Metropolitan Denver, Colorado (CHD). It can also be observed that the genotype frequencies are closer to Luhya, in Webuye, Kenya (LWK) and Maasai, in Kinyawa, Kenya (MKK) populations. An available review including previous studies that investigated the association of several known leptin and leptin receptor variants with obesity29 reported that the results of the included studies were inconclusive. This review shows that the LEPR rs1137100 SNP-obesity association was investigated in five studies and that in one them, the LEPR rs1137100 was associated with obesity in children but not in adults.30 Another study revealed that the LEPR rs1137100 is associated with obesity only in the presence of other two SNPs (LEPR rs8179183 (K656N) and SNP in the LEP gene 3’ flanking region).31 The other three studies related to LEPR rs1137100 included in the review showed no association with obesity or BMI.32–34 The results of the present investigation have shown that the percentage of normal-weight participants carrying the AA genotype is very close to the percentage of overweight and obese participants with the same genotype. In addition, the percentage of the normal-weight participants with the GA or GG genotype is also very close to that of the overweight and obese participants with the same genotype. Consequently, the current study revealed that there is no association between the LEPR rs1137100 and obesity under all the considered genetic models of inheritance. This result is in line with two studies that were conducted in 2011. A systematic review conducted by Bender et al.,34 evaluated the association of several common variants in LEPR including the rs1137100 with overweight while other systematic review and meta-analysis, including seven studies (five European and two Japanese), and enrolling 1,595 cases and 1,173 controls, also showed that there is no significant association of this SNP with obesity susceptibility.25
The mean BMI of participants not carrying any of the LEPR rs1137100 risk alleles (AA) was 31.0, while for those with one risk allele was (GA) the mean BMI was 29.5. For the participants carrying two risk alleles (GG) the determined BMI was 28.7, which is the lowest value. This result contrasts with what was expected, as the BMI decreased in those with both risk alleles. No changes were observed when analyzing the subgroups of participants (men and women, separately). This suggests that the G allele in the LEPR rs1137100 may have a protective effect against increased BMI, which is consistent with a Saudi study conducted in Jeddah city.35 However, they report a significant association with BMI in females but not in males, which contradicts the current study in which the BMI difference determined was not statistically significant. This lack of association with BMI is in line with the results of several other available studies.32,33,36
While most studies have investigated the LEPR rs1137100-obesity association (Table 3), few studies that have analyzed the relationship of this SNP with serum lipid levels. A Japanese study carried out with 136 children reported a significant association between homozygous GG genotype and low levels of total cholesterol and low-density lipoprotein (LDL) cholesterol, but no association was found with triglycerides (TG) and high-density lipoprotein (HD) cholesterol.27 On the contrary, the current study showed a marginal significant association between the LEPR rs1137100 with total and LDL cholesterol, with the GG genotype being associated with higher total and LDL cholesterol levels. However, both studies agree that no association was observed with TG and HDL.
The findings of this study provide valuable insights into the potential role of the rs2167270 variant in the LEP gene as a contributing factor to obesity and metabolic parameters. The results indicate that under the dominant model, the rs2167270 variant is associated with an increased risk of obesity, as evidenced by higher BMI levels. This observation is consistent with previous research that has linked the LEP gene to obesity susceptibility.23 Moreover, the presence of the rs2167270 variant is associated with elevated blood glucose levels and LDL-cholesterol, highlighting its impact on glucose metabolism and lipid regulation. These findings align with other studies that have explored the influence of genetic variants on glucose metabolism and lipid profiles.23
Of note, while the rs2167270 variant showed associations with BMI, blood glucose, and LDL-C, it did not exhibit significant correlations with total cholesterol, HDL-cholesterol, or triglyceride levels. This discrepancy might be attributed to the complex interplay of multiple genetic and environmental factors influencing lipid metabolism.23 Further investigations involving larger and more diverse populations are warranted to comprehensively elucidate the specific genetic determinants of these lipid parameters.
Overall, this study highlights the potential significance of the rs2167270 variant in the LEP gene as a genetic marker for obesity risk and metabolic disturbances. Understanding the role of this genetic variant may aid in the development of targeted interventions and personalized approaches for the management and prevention of obesity-related complications.
The results of the current study suggest that the SNPs; rs2167270, in LEP gene, whereas rs1137100, in the LEPR gene, may not contribute to obesity risk and/or one or more of its related anthropometric and metabolic characteristics in the Jordanian population. This study suggests that the rs2167270 in the LEP gene may contribute to the risk of obesity, under the dominant model, and to higher blood glucose levels, BMI and LDL-C but it does not contribute to the higher total cholesterol, HDL-C or triglycerides. This study also revealed that the rs1137100 in LEPR gene may contribute to the higher total cholesterol and LDL-C blood levels, but that it is not associated with the risk of obesity (evaluated by the BMI value), HDL-C, triglycerides or blood glucose levels. No associations of both studied SNPs with weight or height were found in this study.
The limitations of the study include a small sample size, which limits the statistical power. To confirm the findings presented, studies involving larger Jordanian cohorts should be conducted.
Figshare: Participants Data F1000Research.xlsx, https://doi.org/10.6084/M9.FIGSHARE.25284637.v1. 37
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Obesity, adrenal, thyroid, and diabetes
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Human Genetics, Genetic Association of Metabolic disorders, Genetic structure of Arab populations
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 11 Jun 24 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)