Keywords
CHCHD10, amyotrophic lateral sclerosis, frontotemporal dementia, mitochondrial proteases
CHCHD10 is a small mitochondrial protein with a C-terminal coiled-coil-helix–coiled-coil-helix domain. The N-terminal region of CHCHD10 is mostly intrinsically disordered. Therefore, CHCHD10 has no catalytic activity other than protein-protein interactions through the CHCH domain or intrinsically disordered region. The S59L mutation in CHCHD10 has been identified as a genetic cause of amyotrophic lateral sclerosis and frontotemporal dementia. However, the disease-causing mechanisms of CHCHD10S59L are not yet fully understood. Our previous study showed that CHCHD10S59L induces PINK1 and Parkin accumulation. PINK1 stabilization in the mitochondria is dependent on proteolytic processing by mitochondrial proteases. It has also been reported that mutant CHCHD10 disrupts protein homeostasis within mitochondria. Therefore, we hypothesized that CHCHD10S59L physically interacts with mitochondrial proteases during the disease pathogenesis.
To test this hypothesis, we performed co-immunoprecipitation using transiently transfected wild-type and mutant S59L CHCHD10 in HeLa cells. We employed a dithiobis (succinimidyl propionate) cross-linker to detect transient physical interactions.
We expected that CHCHD10S59L may activate mitochondrial protease processing or expression and that it interacts with more diverse mitochondrial proteases than the wild type. In general, however, there was no difference in the expression and processing levels of mitochondrial proteases and the proteases that interact with wild-type and CHCHD10S59L. LonP1 degrading unfolded proteins in the mitochondrial matrix was the major proteases that can bind to wild-type CHCHD10 and CHCHD10S59L. LonP1 and HtrA2 were significantly less strongly bound to CHCHD10S59L.
This result can be interpreted in many different ways, including the regulatory role of CHCHD10 in mitochondrial proteases. However, more precise biochemical and cell biological investigations are required to determine the role of CHCHD10 in the activity of mitochondrial proteases, mechanism of PINK1 stabilization, and disruption of mitochondrial proteostasis.
CHCHD10, amyotrophic lateral sclerosis, frontotemporal dementia, mitochondrial proteases
Mutations in the coiled-coil-helix-coiled-coil-helix domain 10 (CHCHD10) gene have been identified as the cause of various neuromuscular degenerative diseases, including amyotrophic lateral sclerosis and frontotemporal dementia (ALS-FTD), mitochondrial myopathy, and congenital cardiomyopathy (Ajroud-Driss et al., 2014; Bannwarth et al., 2014). CHCHD10 is a small mitochondrial protein with a C-terminal coiled-coil-helix–coiled-coil-helix domain. The N-terminus of CHCHD10 is mostly intrinsically disordered and contains a hydrophobic region. CHCHD10 has no catalytic activity other than protein-protein interactions through the CHCH domain or intrinsically disordered region (Baek et al., 2021). Among disease-causing mutations, the S59L mutation in the hydrophobic region associated with ALS-FTD has been relatively well characterized in various models. It shows prominent abnormal phenotypes, even in simple cell models such as HeLa or SH-SY5Y cells. Transiently expressed CHCHD10S59L induces mitochondrial fragmentation, reduces ATP production, and reduces oxygen consumption (Baek et al., 2021). The PINK1/Parkin pathway can be activated by CHCHD10S59L protein in both Drosophila and human cells in vivo. Reducing the PINK1/Parkin pathway ameliorates CHCHD10S59L-mediated degeneration and abnormal mitochondrial phenotypes in Drosophila and human cell models (Baek et al., 2021).
PINK1 and Parkin are critical regulators of mitochondrial quality control. PINK1 acts as a sensor of mitochondrial damage, and Parkin promotes the removal of damaged mitochondria through mitophagy. Upon mitochondrial damage, such as loss of mitochondrial membrane potential, PINK1 accumulates in the mitochondria, and this process is dependent on mitochondrial proteasome-mediated PINK1 processing (Greene et al., 2011). Interestingly, however, we did not observe any significant reduction in membrane potential in CHCHD10S59L-transfected HeLa cells showing PINK1 and Parkin accumulation (Baek et al., 2019, 2021), which implies that there may be another factor causing abnormal PINK1 accumulation in CHCHD10S59L-transfected HeLa cells. In addition, it has been reported that mutations in CHCHD10 induced mitochondrial unfolded protein responses and disrupt protein homeostasis within the mitochondria (Genin et al., 2019; Sayles et al., 2022). Taken together, we hypothesized that the expression or activity of mitochondrial proteases may be affected by mutations in CHCHD10, and this change can be achieved by the direct physical interaction between CHCHD10 and mitochondrial proteases. To test this hypothesis, we performed co-immunoprecipitation experiments with a cross-linker, dithiobis (succinimidyl propionate), to retrieve even weak physical interactions between CHCHD10 and the interacting proteins.
All complementary DNAs (cDNAs) for human wild-type and CHCHD10S59L were synthesized and inserted into the pcDNA3.1, vector containing a FLAG tag, by GenScript Inc.
The HeLa cell line was purchased from the American Type Culture Collection (ATCC; CRM-CCL-2). Cells were maintained in Dulbecco’s modified Eagle’s medium (GenClone) supplemented with 10% fetal bovine serum (GenClone), 100 U/mL penicillin and streptomycin (Penicillin-streptomycin, Invitrogen 15140122), and 1 × GlutaMax (Gibco 35050061). Cells were transfected using the jetPRIME (Polyplus) transfection reagent according to the manufacturer’s protocol with an automatic cell counter (NanoEntek, ADAM-MC2 cell counter).
Dithiobis (succinimidyl propionate) (DSP, Lomant’s reagent) was purchased from Thermo Fisher Scientific (22586) and used to induce protein cross-linking. HeLa cells transfected with FLAG-tagged CHCHD10 WT or S59L were washed twice with PBS to remove the medium. After adding PBS containing a final concentration of 2 mM DSP, the cells were incubated on ice for 2 h. The reaction was stopped by adding 1M Tris, pH 7.5 (at a final concentration of 20 mM) and incubated for 15 min. Cells were washed twice with PBS and solubilized with NP-40 lysis buffer (20 mM Tris, 137 mM NaCl, 1% NP-40, and 2 mM EDTA with a protein inhibitor cocktail). After sonication on ice, lysates were collected by centrifugation. Equal amounts of protein lysates were incubated with beads conjugated with anti-FLAG antibodies (Sigma, ANTI-FLAG® M2 Affinity Gel, A2220) overnight at 4 °C. After washing thrice with NP-40 lysis buffer, the pellets were resuspended in 1× reduced LDS sample buffer. After heating the samples at 70 °C for 10 min, the samples were subjected to SDS-PAGE and immunoblotting. The following primary antibodies were used for immunoblotting: FLAG (Sigma F1804 mouse monoclonal, and Proteintech 20543-1-AP rabbit polyclonal, 1:1000), AFG3L2 (Proteintech 14631-1-AP rabbit polyclonal, 1:1000), PARL (Proteintech 26679-1-AP rabbit polyclonal, 1:1000), LonP1 (Proteintech 15440-1-AP rabbit polyclonal, 1:1000), PMPCA (Proteintech 26536-1-AP rabbit polyclonal 1:1000), HTRA2 (Proteintech 15775-1-AP rabbit polyclonal, 1:1000), CLPP (Proteintech 15698-1-AP rabbit polyclonal, 1:1000), PITRM1 (Proteintech 26536-1-AP rabbit polyclonal, 1:1000), and actin (Proteintech 20536-1-AP rabbit polyclonal, Santa Cruz Biotechnology SC-47778 Mouse monoclonal, 1:5000). Immunoblots were visualized and analyzed using a fluorescence imaging system (LI-COR, Odyssey® FX Imager).
Graphs for the quantitative analysis of immunoblots (mean difference between groups with 95% confidence interval) were generated using Estimation Stats (Ho et al., 2019) (http://estimationstats.com). We performed an unpaired two-sided permutation t-test to evaluate the data with p-values.
We examined seven mitochondrial proteases for their ability to interact with wild-type CHCHD10 or CHCHD10S59L using transiently transfected HeLa cells and DSP treatment (Figure 1A). The proteases were selected based on information from BioGRID for CHCHD10 and its paralog and binding partner, CHCHD2, as well as the literature on PINK1 processing proteases (Greene et al., 2011). Intact cells were treated with DSP and washed before cell lysis to prevent cross-linking post-lysis. Initially, we examined whether there were any significant changes in protein expression for all seven proteases by wild-type or CHCHD10S59L expression. Although there were some variations among repeated experiments, such as Presenilin-Associated Rhomboid-Like protein (PARL, see supplementary file), we did not observe any significant and consistent changes in the expression level or processing of proteases due to the transient expression of wild-type or CHCHD10 S59L. PARL is involved in the proteolytic cleavage of important substrates, such as OPA1 and PINK1, for maintaining mitochondrial morphology and function (Jin et al., 2010). However, PARL did not interact with CHCHD10, contrary to our hypothesis. Another protease that may be related to PINK1 processing as a component of ClpXp complexes (Greene et al., 2011), CLPP, also did not co-immunoprecipitate with CHCHD10 using an anti-FLAG antibody, although a component of ClpXp, ClpX, was enlisted in BioGRID as an interacting partner of CHCHD10 or CHCHD2. In addition, PITRM1 (Pitrilysin Metallopeptidase 1) degrading cleaved mitochondrial targeting sequences (MTS), and other small peptides (Ståhl et al., 2002) have also been reported to interact with both CHCHD10 and CHCHD2. However, we were unable to detect PITRM1 in our repeated co-immunoprecipitation experiments.
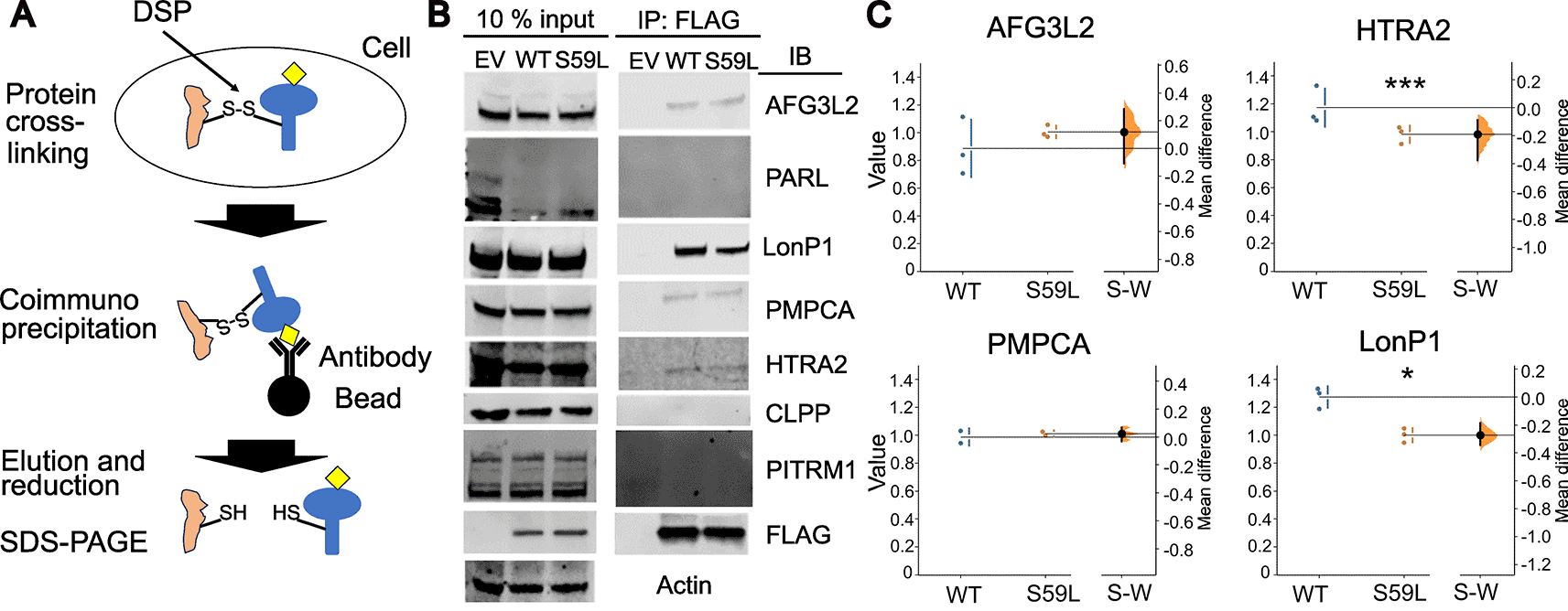
(A) Schematic diagram of in-cell DSP cross-linking and immunoprecipitation with anti-FALG antibody (see methods for a detailed procedure), (B) western blot results with mitochondrial proteases antibodies, 10% of total lysates were used in the left panel, and co-immunoprecipitated proteases were detected in the right panel, (C) quantification of interacting proteases. The raw data are plotted on the right axes. On the left axes, mean differences are plotted. Each 95% confidence interval is indicated by the ends of the vertical error bars. S-W: the difference between S59L and wild-type groups. * <0.05 and *** <0.001.
AFG3L2 is a mitochondrial AAA protease that can process PINK1. It has been reported that siRNA treatment for AFG3L2 causes PINK1 accumulation (Thomas et al., 2014). PMPCA is a subunit of MPP that initially processes PINK1(Greene et al., 2011). Both proteases interact with wild-type CHCHD10 and CHCHD10S59L. However, their interactions were weak, and there were no significant differential interactions, although AFG3L2 tended to bind more to mutant S59L CHCHD10. HTRA2 is a member of the high-temperature requirement A family of proteases that bind to PINK1 (Plun-Favreau et al., 2007). Evidence suggests that HTRA2 is associated with Parkinson’s disease (Plun-Favreau et al., 2007). Notably, the S59L mutation in CHCHD10 resulted in a decreased interaction between HTRA2 and CHCHD10 compared to binding with wild-type CHCHD10. The results were statistically significant. However, their interaction was the weakest among the seven proteases, and the physiological significance of this interaction is uncertain.
The protease that most significantly interacted with CHCHD10 was LonP1. LonP1 is not listed in BioGRID for CHCHD10 or CHCHD2. Both wild-type and CHCHD10S59L interacted significantly with LonP1. It has been reported that various stress conditions, including reactive oxygen species and unfolded protein stress, can upregulate LonP1 expression. However, we did not observe any significant changes in the expression levels in transiently transfected wild-type CHCHD10 or mutant S59L CHCHD10. LonP1 also degrades PINK1, and silencing LonP1 enhances PINK1 accumulation with subsequent activation of the PINK1-Parkin mitophagy pathway (Jin & Youle, 2013; Thomas et al., 2014). In our co-immunoprecipitation results, the interaction between LonP1 and CHCHD10 was significantly decreased by the S59L mutation.
By co-immunoprecipitation experiments with the cross-linker DSP, we determined that PARL, CLPP, and PITRM1 do not interact with wild-type or mutant S59L CHCHD10. AFG3L2 and PMPCA interacted with both CHCHD10 and mutant S59L CHCHD10. However, no differential interactions were caused by the S59L mutation. Interestingly, two proteases, HTRA2 and LonP1, bind to CHCHD10, and the S59L mutation decreased their interaction. Since we did not observe any consistent expression or processing changes caused by transiently transfected CHCHD10, the changes in the physical interaction between CHCHD10 and mitochondrial proteases may be related to protease activity. The decreased interaction between mutant S59L CHCHD10 and HTRA2 and LonP1 implies that wild-type CHCHD10 somehow maintains the active state of HTRA2 and LonP1, and the mutant S59L protein loses this activity partially due to their decreased interaction. Therefore, less active HTRA2 and LonP1 could not perform their roles in degrading unfolded proteins, preventing protein aggregation, and processing PINK1. However, the role of the physical interaction between CHCHD10 and mitochondrial proteases should be determined by further biochemical analyses, and the effect of the S59L mutation should also be precisely determined.
Differential physical interactions of wild-type and S59L mutant CHCHD10 with mitochondrial proteases.
Figshare: Co-IP with CHCHD10 (https://doi.org/10.6084/m9.figshare.26090764) (Kim and Oh, 2024).
This project contains the following underlying data:
Statistics data analysis (chchd10 interaction partner).xlsx (raw data for estimation statistics analysis)
Supple.zip (raw image files for all western blots)
Suppl.pptx (individual blots with annotations used in Figure 1)
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Neurodegenerative diseases including ALS/FTD and protein-protein interactions.
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Neurodegeneration and biochemical assays
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Biochemistry and cell biology on mitochondria.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 02 Jul 24 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)