Keywords
Mirtron, Secondary Structure
Mirtrons, a vital category of non-canonical microRNAs (miRNAs) originating from exon-intron boundaries through splicing mechanisms, play crucial roles in cellular processes. However, existing databases lack the latest data and structural information, hindering understandings of mirtron formation and functions.
We introduce MirtronStructDB, an online database addressing these gaps by incorporating over 350 novel mirtrons. Significantly, it provides corresponding predicted RNA secondary structures, offering a deeper understanding of the functional roles and mechanisms of all mirtrons. This enhances previous repositories, offering a total of 4,209 mirtron records spanning 25 species from 46 publications. Our database contributes for unraveling patterns and functions in mirtrons across species and diverse structural features. MirtronStructDB allows users to freely browse, search, visualize, and download data via a user-friendly interface.
MirtronStructDB is accessible at: http://www.bio8.cs.hku.hk/msdb/.
lishumin@connect.hku.hk, rbluo@cs.hku.hk
Mirtron, Secondary Structure
Mirtrons are a vital category of non-canonical microRNAs (miRNAs). MicroRNAs represent a crucial class of small RNAs that play significant roles in various post-transcriptional cellular processes.1 Conventionally, miRNAs are processed from longer primary transcripts, known as pri-miRNAs, via sequential enzymatic cleavage steps involving the Drosha or Dicer proteins. However, a distinct class of miRNAs, termed mirtrons, have been identified that they follow an alternative processing pathway. Mirtrons originate from exon-intron boundaries through splicing and are independent of Drosha or Dicer cleavage. The initial detection of mirtrons occurred in D. melanogaster and C. elegans2 and subsequent studies confirmed their presence in mammals,3 plants,4 and viruses.5 Notably, mirtrons have demonstrated involvement in diverse biological processes, including cell development and cancer.6 To facilitate the exploration of mirtrons, an organized database, MirtronDB, was launched in 2019, collecting 3,833 precursors or mature mirtrons across 18 species.7 However, MirtronDB is limited by the absence of the latest data and, more significantly, structural information—a critical factor in miRNA maturation and distinguishing canonical and non-canonical miRNAs.8
To address these limitations, we present MirtronStructDB, an updated database that offers corresponding secondary structures of all mirtrons, along with an addition of over 350 novel mirtrons that are currently unavailable in existing databases. Furthermore, our user-friendly web interface provides browsing, searching, visualization, and downloading of all stored data. Through timely updates and comprehensive structural information, MirtronStructDB aims to contribute to the advancement of mirtron research and facilitate investigations into their functional roles and mechanisms. MirtronStructDB is publicly accessible at: http://www.bio8.cs.hku.hk/msdb/.
Data collection and processing
MirtronStructDB was constructed through a two-fold data collection strategy (Figure 1A). Initially, we obtained the data from mirtronDB7 by downloading their available information as a primary source. Subsequently, we conducted a comprehensive literature search using the term ‘mirtron OR mirtrons’ from PubMed to collect additional publications related to mirtron discoveries. All the collected data underwent a standardization process and was integrated into MirtronStructDB. In cases where certain fields were missing from the original papers, temporarily placeholders (‘tbu’ indicating ‘to be updated’) were employed. To augment the database and investigate the functions and mechanisms of diverse mirtrons, we predicted the RNA secondary structures for all the collected data with sequence information. This prediction was performed through the RNAstructure Web Server with default parameters.9
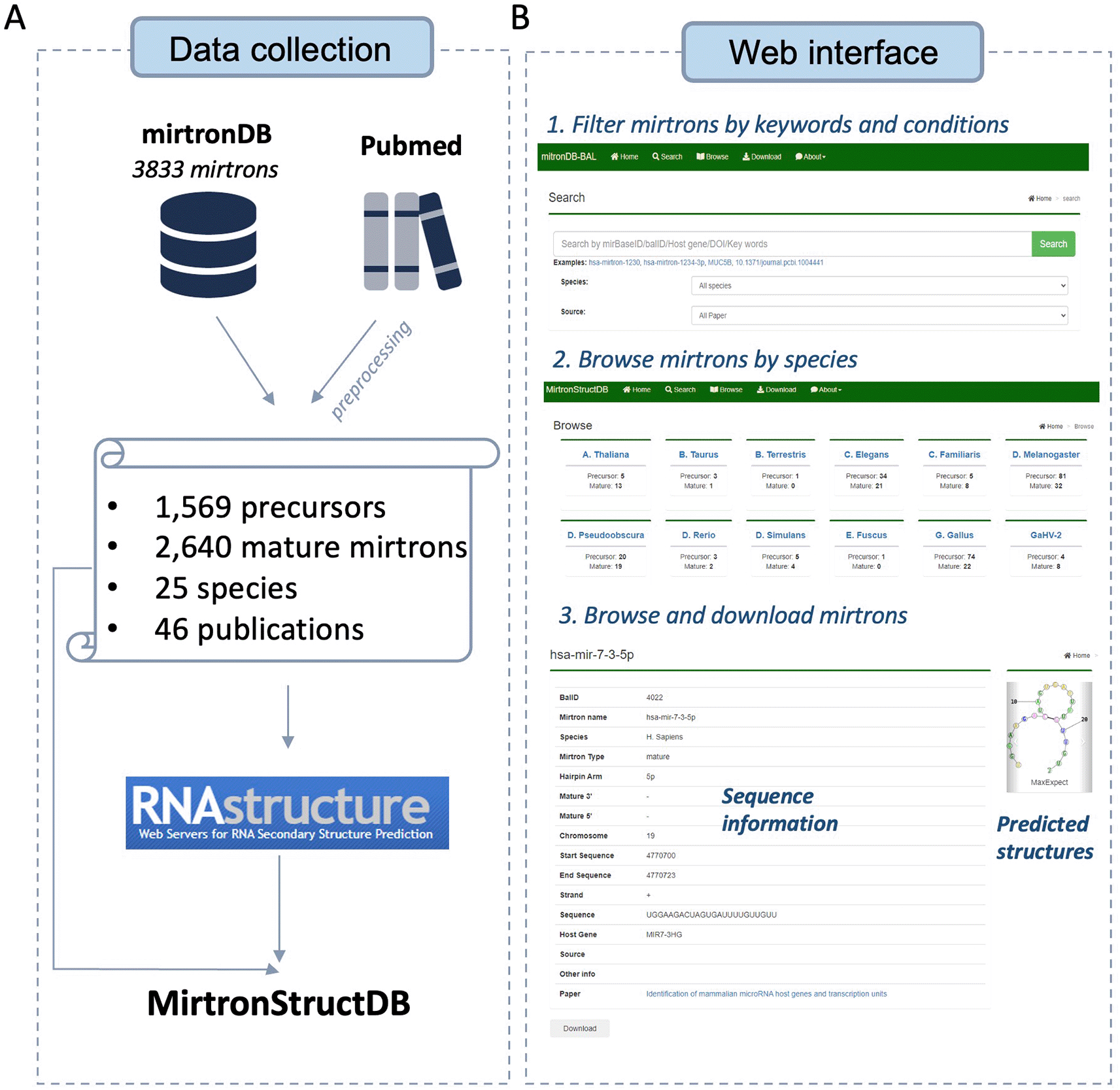
A) Workflow of data collection and preprocessing. B) Main modules of MirtronStructDB: search and filter, browse by species, demonstration and download.
In summary, MirtronStructDB offers a comprehensive dataset comprising 1,569 precursors and 2,640 mature mirtrons spanning 25 species, sourced from 46 publications. Of these, 163 precursors and 223 mature mirtrons were first documented in a dedicated mirtron database. And 9 species were first collected and reported in MirtronStructDB. In addition, a total of 12,555 RNA secondary structures were predicted for better understandings of the functional roles and mechanisms of mirtrons. Detailed information was provided in Data availability statement.
Web application implementation
The online web server was developed with the Flask web framework (v1.1.4) (https://github.com/pallets/flask) as the backend. It is deployed on an Ubuntu Linux server equipped with 48 Intel Xeon CPUs and 189GB of memory. All data is stored in a SQLite database, managed by the SQLAlchemy toolkit (v2.5.1) (https://www.sqlalchemy.org/).
The frontend of MirtronStructDB with a user-friendly design, crafted using Bootstrap (v3) (https://getbootstrap.com/) and adminLTE (v2.4.18) (https://github.com/ColorlibHQ/AdminLTE), and dynamically generated through the JINJA2 templating engine (v2.11.3) (https://jinja.palletsprojects.com/en/3.1.x/templates/). The web service is hosted by Apache2 and Gunicore modules, guaranteeing high performance and stability.
MirtronStructDB is available to the community through its web-based interface and is freely available. It can also be run locally with a typically 2GB RAM and a dual-core CPU. It is compatible with popular web browsers, including Microsoft Edge, Google Chrome, Firefox and Safari.
The web portal has three primary modules: search and filter, browse by species, and demonstration and download. The overview is shown in Figure 1B. Users can either start with search and filter or browse by species to get the mirtron list and then select individual mirtrons to view their details. Usages listed below:
Search and filter: To facilitate the use of MirtronStructDB, a search and filter can be performed through clicking the ‘Search’ button from the top navigation bar. Users can type the IDs of mirBase, host gene symbols, DOIs or keywords of papers in the search box to retrieve all relevant mirtrons. Additionally, users can apply filters based on preferred species and sources to customize their search results.
Browse by species: We also provide a gallery page which categorizing mirtrons by species. Users can access it through clicking ‘Browse’ button of the navigation bar. Mirtron summaries for each species are presented, and users can explore detailed information by clicking on individual species.
Demonstration and download: Individual mirtrons can be accessed through the mirtron detail page. Two sections were shown: The first section includes basic information including the species, corresponding precursors or mature mirtrons, sequences, strand, chromosome position, source papers, etc. The second section displays predicted secondary structures. Users are flexible to explore and download data based on their own needs.
MirtronStructDB is a comprehensive database with an extensive collection of over 350 novel mirtrons accompanied by their predicted secondary structures. This database not only presents an updated catalog of mirtrons across diverse species but also provide structural insights into the entire spectrum of mirtrons. MirtronStructDB make it feasible for experimental biologists seeking identification of relevant mirtrons for distinct species, sources, or specific research requirements. Furthermore, MirtronStructDB offers computational biologists an opportunity to delve into its rich information, particularly the sequence contexts and structural features it provides. With the accumulated data of mirtronStructDB, we anticipate the emergence of novel computational methods to expedite the prediction and identification of mirtrons across a broader spectrum of species. This advancement is expected to deepen our understanding of the unique formation mechanisms and functions of mirtrons.
R. L. And S. L. conceived the study. S. L. designed the web interface and the analyses. F. W. C., L. C., J. S., and S. S. A. evaluated the analysis results. All authored drafted and approved the manuscript.
Zenodo: Source data of Mirtronstructdb - A comprehensive database of mirtrons with predicted secondary structure. https://zenodo.org/doi/10.5281/zenodo.13118506. 10
The project contains the following underlying data:
- msdb_data.csv (the mirtron collection table, including the following columns: Unique ID, mirtron name, type, species, hairpin arm, sequence, chromosome, start, end, strand, host gene, 3p-arm mature miRNAs, 5p-arm mature miRNAs, precursor name, data source, PMID, paper title, DOI, other information).
- msdb_structures.tar.gz (the SVG images of the predicted secondary structures based on the mirtron sequences).
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Database available from: http://www.bio8.cs.hku.hk/msdb/ Source code available from: https://github.com/HKU-BAL/msdb-flask/.
Archived software available from https://zenodo.org/doi/10.5281/zenodo.13118891. 11
License: MIT
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
No
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
No
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bioinformatics, genomics and transcriptomics
Is the rationale for developing the new software tool clearly explained?
Partly
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Structural Bioinformatics, RNAi
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 06 Aug 24 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)