Keywords
UniProt ID P42858, HTT, Huntingtin, antibody characterization, antibody validation, western blot, immunoprecipitation, immunofluorescence
This article is included in the YCharOS (Antibody Characterization through Open Science) gateway.
Huntingtin encodes a 3144 amino acid protein, with a polyglutamine repeat tract at the N-terminus. Expansion of this repeat tract above a pathogenic threshold of 36 repeats is the causative mutation of Huntington's disease, a neurodegenerative disorder characterized by loss of striatal neurons. Here we have characterized twenty Huntingtin commercial antibodies for western blot, immunoprecipitation, and immunofluorescence using a standardized experimental protocol based on comparing read-outs in knockout cell lines and isogenic parental controls. These studies are part of a larger, collaborative initiative seeking to address antibody reproducibility issues by characterizing commercially available antibodies for human proteins and publishing the results openly as a resource for the scientific community. While use of antibodies and protocols vary between laboratories, we encourage readers to use this report as a guide to select the most appropriate antibodies for their specific needs.
UniProt ID P42858, HTT, Huntingtin, antibody characterization, antibody validation, western blot, immunoprecipitation, immunofluorescence
An error occurred as only half of the antibodies tested by our team were included in Figure 3. The original submission featured a split Figure 3 with Parts 1 and 2 uploaded separately. The revised Figure 3 now includes all 20 HTT antibodies tested.
To read any peer review reports and author responses for this article, follow the "read" links in the Open Peer Review table.
Huntington’s Disease (HD) is a neurodegenerative disorder inherited in an autosomal dominant manner, presenting with a spectrum of progressive motor, cognitive, and psychological impairments, typically with adult-onset of symptoms.1 Although the HD causative gene, HTT, was discovered over three decades ago, there are still no disease-modifying treatments available for patients, and progress unpicking the molecular pathology of the disease remains slow.2
HD arises from a heterozygous expansion mutation of the trinucleotide CAG repeat tract in exon 1 of HTT, located on chromosome 4, above a critical threshold of ~36 repeats. This mutation results in expansion of the polyglutamine stretch at the N-terminus of the 3144 amino acid Huntingtin protein. Huntingtin functions as a scaffold protein, engaging in extensive protein-protein interactions,3 forming various multi-protein complexes to carry-out its diverse array of functions. Modulation of this interaction network by the polyglutamine expansion contributes to degeneration within the central nervous system, affecting medium spiny neurons at the onset of disease.4,5
The low expression level, complex interactome and large size of the 348 kDa Huntingtin protein have given rise to technical challenges which have hindered precise determination of its molecular function, or how this is altered in disease. In particular, the use of different Huntingtin antibodies by scientists in the HD research community, often mapping to structurally distant epitopes, can yield different or even conflicting results, further conflating interrogation of this protein.6–8
This research is part of a broader collaborative initiative in which academics, funders and commercial antibody manufacturers are working together to address antibody reproducibility issues by characterizing commercial antibodies for human proteins using standardized protocols, and openly sharing the data.9–11 Here we evaluated the performance of twenty commercial antibodies for Huntingtin for use in western blot, immunoprecipitation, and immunofluorescence, enabling biochemical and cellular assessment of Huntingtin properties and function. The platform for antibody characterization used to carry out this study was endorsed by a committee of industry and academic representatives. It consists of identifying human cell lines with adequate target protein expression and the development/contribution of equivalent knockout (KO) cell lines, followed by antibody characterization procedures using most commercially available antibodies against the corresponding target protein. The standardized consensus antibody characterization protocols are openly available on Protocol Exchange (DOI: 10.21203/rs.3.pex-2607/v1).12
The authors do not engage in result analysis or offer explicit antibody recommendations. A limitation of this study is the use of universal protocols - any conclusions remain relevant within the confines of the experimental setup and cell line used in this study. Our primary aim is to deliver top-tier data to the scientific community, grounded in Open Science principles. This empowers experts to interpret the characterization data independently, enabling them to make informed choices regarding the most suitable antibodies for their specific experimental needs. Guidelines on how to interpret antibody characterization data found in this study are featured on the YCharOS gateway.13
Our standard protocol involves comparing readouts from WT (wild type) and KO cells.14,15 The first step is to identify a cell line(s) that expresses sufficient levels of a given protein to generate a measurable signal. To this end, we examined the DepMap transcriptomics database to identify all cell lines that express the target at levels greater than 2.5 log2 (transcripts per million “TPM” + 1), which we have found to be a suitable cut-off (Cancer Dependency Map Portal, RRID:SCR_017655). We generated an HTT KO line in DMS 53 as it expresses the endogenous HTT transcript at 6.1 log2 (TPM+1), which is above the average range of cancer cell lines analyzed. A commercial HAP1 HTT KO is also available; HAP1 expresses HTT at 3.7 log2 (TPM+1) RNA level. A HEK293T HTT KO cell line has been developed and used elsewhere16 ( Table 1). All three cell line backgrounds were evaluated by western blot using a high-performing Huntingtin antibody detected in Figure 1. DMS 53 was identified as the most suitable cell line ( Figure 2), which can be explained by its high expression of the HTT transcript compared to other two cell lines. Thus, DMS 53 WT and KO cell lines were generated and used to evaluate the antibodies in all applications.
| Institution | Catalog number | RRID (Cellosaurus) | Cell line | Genotype |
|---|---|---|---|---|
| ATCC | CRL-2062 | CVCL_1177 | DMS 53 | WT |
| Academic | non-commercial | CVCL_D6U0 | DMS 53 | HTT KO |
| ATCC | CRL-3216 | CVCL_0063 | HEK 293T | WT |
| Academic | non-commercial | CVCL_D7EP | HEK 293T | HTT KO16 |
| Horizon Discovery | C631 | CVCL_Y019 | HAP1 | WT |
| Horizon Discovery | HZGHC004595c006 | CVCL_SR86 | HAP1 | HTT KO |
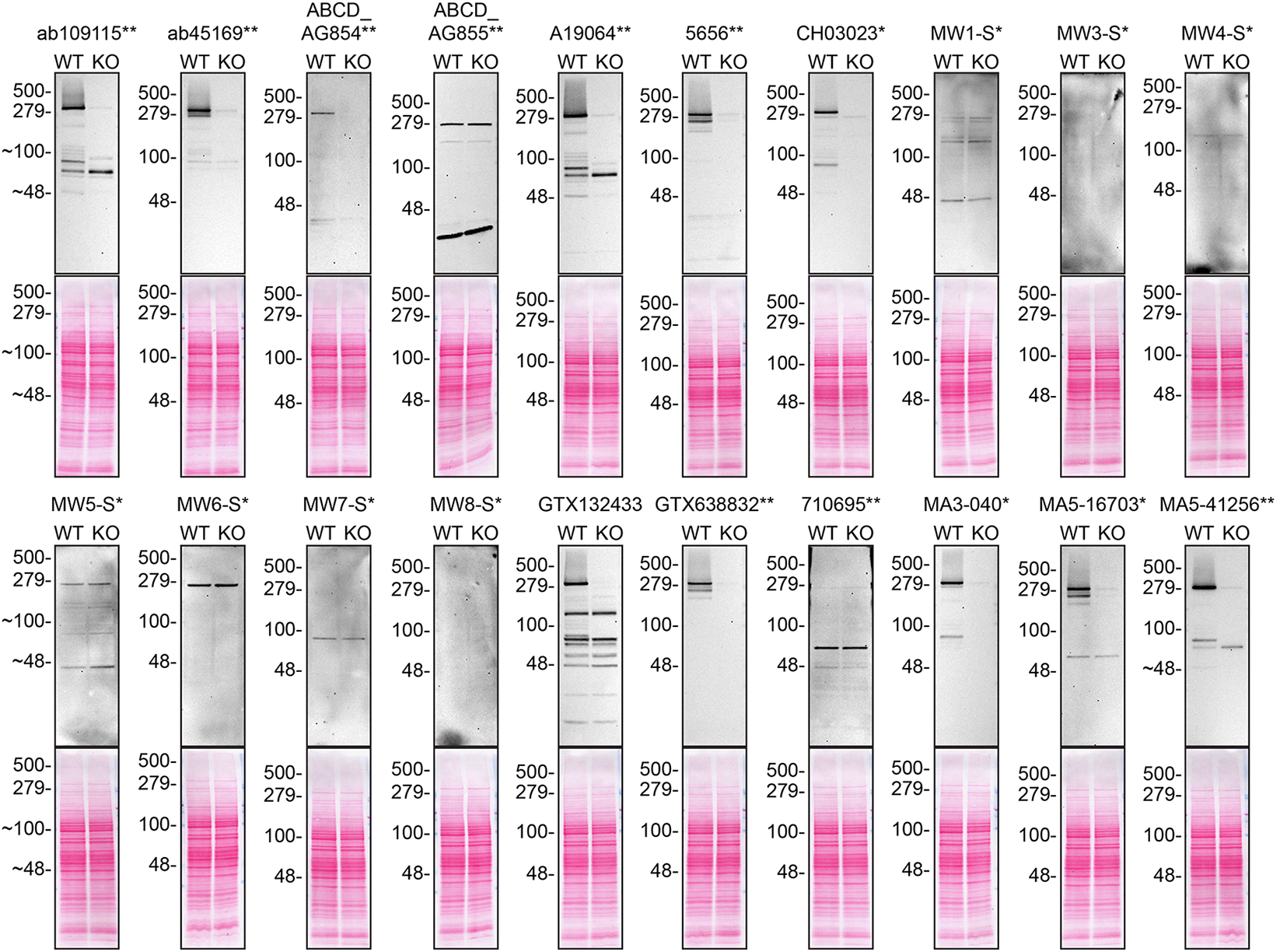
Lysates of DMS 53 (WT and HTT KO) were prepared and 30 μg of protein were processed for western blot with the indicated Huntingtin antibodies. Tris-Glycine 4-20% gels were used for SDS-PAGE. The Ponceau stained transfers of each blot are presented to show equal loading of WT and KO lysates and protein transfer efficiency from the acrylamide gels to the nitrocellulose membrane. Antibody dilutions were chosen according to the recommendations of the antibody supplier. Exceptions were given for antibodies ab109115**, ab45169**, and MA5-41256** which were titrated as the signal was too weak when following the supplier’s recommendations. Antibody dilution used: ab109115** at 1/2000, ab45169** at 1/5000, ABCD_AG854** at 1/10, ABCD_AG855** at 1/10, A19064** at 1/1000, 5656** at 1/1000, CH03023* at 1/1000, MW1-S* at 1/10, MW3-S* at 1/10, MW4-S* at 1/10, MW5-S* at 1/10, MW6-S* at 1/10, MW7-S* at 1/10, MW8-S* at 1/10, GTX132433 at 1/500, GTX638832** at 1/500, 710695** at 1/200, MA3-040* at 1/1000, MA5-16703* at 1/500, MA5-41256** at 1/2000. Predicted band size: 347 kDa. *Monoclonal antibody, **Recombinant antibody.
Note: MW1-S*, MW3-S*, MW4-S*, MW5-S*, MW6-S*, MW7-S* and MW8-S* are expected to only recognize an altered conformation of the polyQ domain generated as the polyQ domain of Huntingtin increases in length.

Lysates of WT and HTT KO in DMS 53, HEK 293T and HAP1 were prepared, and 30 μg of protein was processed for western blot with the indicated Huntingtin antibodies ab45169** at 1/5000 and GTX638832** at 1/500. Tris-Glycine 4-20% gels were used for SDS-PAGE. The Ponceau stained transfer is shown as a loading control. Predicted band size: 347 kDa. **Recombinant antibody.
For western blot experiments, WT and HTT KO protein lysates were ran on SDS-PAGE, transferred onto nitrocellulose membranes, and then probed with twenty Huntingtin antibodies in parallel ( Table 2, Figure 1).
| Company | Catalog number | Lot number | RRID (Antibody Registry) | Clonality | Clone ID | Host | Concentration (μg/μl) | Vendors recommended applications |
|---|---|---|---|---|---|---|---|---|
| Abcam | ab109115** | 1003147-4 | AB_10863082 | recombinant-mono | EPR5526 | rabbit | 1.52 | Wb, IF |
| Abcam | ab45169** | 1022500 | AB_733062 | recombinant-mono | EP867Y | rabbit | 1.55 | Wb, IF |
| ABCD Antibodies | ABCD_AG854** | 10/27/2023 | AB_3076339 | recombinant-mono | 12.3 | rabbit | 0.004 | n/a |
| ABCD Antibodies | ABCD_AG855** | 10/27/2023 | AB_3076340 | recombinant-mono | C4 | rabbit | 0.19 | n/a |
| ABclonal | A19064** | 4000000431 | AB_2862557 | recombinant-mono | ARC0431 | rabbit | 0.63 | Wb, IF |
| Cell Signaling Technology | 5656** | 6 | AB_10827977 | recombinant-mono | D7F7 | rabbit | 0.10 | Wb, IF |
| Coriell Institute | CH03023* | 03.17.21 | AB_3096092 | monoclonal | 2B7 | mouse | 1.71 | n/a |
| DSHB | MW1-S* | 44322 | AB_528290 | monoclonal | MW1-S | mouse | 0.02 | Wb, IP, IF |
| DSHB | MW3-S* | 43216 | AB_528292 | monoclonal | MW3-S | mouse | 0.01 | Wb, IF |
| DSHB | MW4-S* | 43251 | AB_528293 | monoclonal | MW4-S | mouse | 0.02 | Wb, IF |
| DSHB | MW5-S* | 44742 | AB_528294 | monoclonal | MW5-S | mouse | 0.03 | Wb, IF |
| DSHB | MW6-S* | 43230 | AB_528295 | monoclonal | MW6-S | mouse | 0.06 | Wb, IF |
| DSHB | MW7-S* | 43461 | AB_528296 | monoclonal | MW7-S | mouse | 0.03 | Wb, IF |
| DSHB | MW8-S* | 44315 | AB_528297 | monoclonal | MW8-S | mouse | 0.04 | Wb, IP, IF |
| GeneTex | GTX132433 | 42312 | AB_2886646 | polyclonal | MW3-S | rabbit | 1.40 | Wb, IF |
| GeneTex | GTX638832** | 45096 | AB_3094813 | recombinant-mono | HL2483 | rabbit | 0.98 | Wb |
| Thermo Fisher Scientific | 710695** | RF236710 | AB_2608784 | recombinant-poly | 3HCLC | rabbit | 0.50 | IF |
| Thermo Fisher Scientific | MA3-040* | YG376237 | AB_2608783 | monoclonal | 1HU-4C8 | mouse | n/a | Wb, IF |
| Thermo Fisher Scientific | MA5-16703* | YE3913821A | AB_2538195 | monoclonal | HDB4E10 | mouse | 1.00 | Wb, IP, IF |
| Thermo Fisher Scientific | MA5-41256** | YE3913382B | AB_2899009 | recombinant-mono | JB89-34 | rabbit | 1.00 | Wb, IF |
We then assessed the capability of all twenty antibodies to capture Huntingtin from DMS 53 protein extracts using immunoprecipitation techniques, followed by western blot analysis. For the immunoblot step, a specific Huntingtin antibody identified previously (refer to Figure 1) was selected. Equal amounts of the starting material (SM), the unbound fraction (UB), as well as the whole immunoprecipitate (IP) eluates were separated by SDS-PAGE ( Figure 3).

DMS 53 lysates were prepared, and immunoprecipitation was performed using 2.0 μg of the indicated Huntingtin antibodies pre-coupled to Dynabeads protein A or protein G. The concentration of MA3-040* is unknown and therefore 5 μL of this antibody was tested. All samples were washed and processed for western blot with the indicated Huntingtin antibody. Tris-Glycine 4-20% gels were used for SDS-PAGE. For western blot, ab45169** was used at 1/5000. The Ponceau stained transfers of each blot are shown. SM=4% starting material; UB=4% unbound fraction; IP=immunoprecipitate. *Monoclonal antibody, **Recombinant antibody.
For immunofluorescence, twenty antibodies were screened using a mosaic strategy. First, DMS 53 WT and HTT KO cells were labelled with different fluorescent dyes in order to distinguish the two cell lines, and the Huntingtin antibodies were evaluated. Both WT and KO lines were imaged in the same field of view to reduce staining, imaging and image analysis bias ( Figure 4). Quantification of immunofluorescence intensity in hundreds of WT and KO cells was performed for each antibody tested,12 and the images presented in Figure 3 are representative of this analysis.

DMS 53 WT and HTT KO cells were labelled with a green or a far-red fluorescent dye, respectively. WT and KO cells were mixed and plated to a 1:1 ratio on coverslips. Cells were stained with the indicated Huntingtin antibodies and with the corresponding Alexa-fluor 555 coupled secondary antibody including DAPI. Acquisition of the blue (nucleus-DAPI), green (WT), red (antibody staining) and far-red (KO) channels was performed. Representative images of the merged blue and red (grayscale) channels are shown. WT and KO cells are outlined with green and magenta dashed line, respectively. When an antibody was recommended for immunofluorescence by the supplier, we tested it at the recommended dilution and at 1 μg/ml. The rest of the antibodies were tested at 1 and 2 μg/ml. The final concentration of each antibody was selected based on the detection range of the microscope used and a quantitative analysis not shown here. Antibody dilutions corresponding to the images shown are: ab109115** at 1/1500, ab45169** at 1/1500, ABCD_AG854** at 1/500, ABCD_AG855** at 1/200, A19064** at 1/600, 5656** at 1/100, CH03023* at 1/1700, MW1-S* at 1/20, MW3-S* at 1/10, MW4-S* at 1/10, MW5-S* at 1/15, MW6-S* at 1/60, MW7-S* at 1/15, MW8-S* at 1/20, GTX132433 at 1/500, GTX638832** at 1/500, 710695** at 1/500, MA3-040* at 1/1000, MA5-16703* at 1/1000, MA5-41256** at 1/1000. Bars = 10 μm. *Monoclonal antibody, **Recombinant antibody.
In conclusion, we have screened twenty commercial Huntingtin antibodies by western blot, immunoprecipitation, and immunofluorescence by comparing the signal produced by the antibodies in human DMS 53 WT and HTT KO cells. Several high-quality and renewable Huntingtin antibodies were identified in all applications. Researchers who wish to study Huntingtin in a different species are encouraged to select high-quality antibodies, based on the results of this study, and investigate the predicted species reactivity of the manufacturer before extending their research.
The underlying data for this study can be found on Zenodo, an open-access repository for which YCharOS has its own collection of antibody characterization reports.17
The standardized protocols used to carry out this KO cell line-based antibody characterization platform was established and approved by a collaborative group of academics, industry researchers and antibody manufacturers. The detailed materials and step-by-step protocols used to characterize antibodies in western blot, immunoprecipitation and immunofluorescence are openly available on Protocol Exchange, a preprint server (DOI: 10.21203/rs.3.pex-2607/v1).12
Cell lines used and primary antibodies tested in this study are listed in Tables 1 and 2, respectively. To ensure that the cell lines and antibodies are cited properly and can be easily identified, we have included their corresponding Research Resource Identifiers, or RRID.18,19
Zenodo: Dataset for the Huntingtin antibody screening study, doi.org/10.5281/zenodo.11639052.17
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
We would like to thank the NeuroSGC/YCharOS/EDDU collaborative group for their important contribution to the creation of an open scientific ecosystem of antibody manufacturers and KO cell line suppliers, for the development of community-agreed protocols, and for their shared ideas, resources, and collaboration. Members of the group can be found below. We would also like to thank the Advanced BioImaging Facility (ABIF) consortium for their image analysis pipeline development and conduction (RRID:SCR_017697). Members of each group can be found below.
NeuroSGC/YCharOS collaborative group: Thomas M. Durcan, Aled M. Edwards, Peter S. McPherson, Chetan Raina and Wolfgang Reintsch.
ABIF consortium: Claire M. Brown and Joel Ryan.
Thank you to the Structural Genomics Consortium, a registered charity (no. 1097737), for your support on this project. The Structural Genomics Consortium receives funding from Bayer AG, Boehringer Ingelheim, Bristol-Myers Squibb, Genentech, Genome Canada through Ontario Genomics Institute (grant no. OGI-196), the EU and EFPIA through the Innovative Medicines Initiative 2 Joint Undertaking (EUbOPEN grant no. 875510), Janssen, Merck KGaA (also known as EMD in Canada and the United States), Pfizer and Takeda.
An earlier version of this of this article can be found on Zenodo (DOI: 10.5281/zenodo.11582780).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: In vivo and post-mortem imaging of neurodegenetive disorders, including HD
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Mechanisms of neurodegenerative disease
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Partly
Are sufficient details of methods and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a useable and accessible format?
Yes
References
1. DiFiglia M, Sapp E, Chase KO, Davies SW, et al.: Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain.Science. 1997; 277 (5334): 1990-3 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Pathogenesis of HD and 3D histology and pathology of human disease.
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a useable and accessible format?
Yes
References
1. Ko J, Ou S, Patterson PH: New anti-huntingtin monoclonal antibodies: implications for huntingtin conformation and its binding proteins.Brain Res Bull. 56 (3-4): 319-29 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: The molecular pathogenesis of Huntingtin's disease, pre-clinical Huntington's disease mouse models, HTT protein bioassays, Molecular biology, HTT protein aggregation.
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: drug discovery and diagnostics
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Cell biology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
|
Version 2 (revision) 02 Jan 25 |
read | read | read | read | ||
|
Version 1 13 Aug 24 |
read | read | ||||
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)