Keywords
Quantum Machine Learning, Quantum Graph Learning, Graph Neural Networks, Quantum Feature Embedding
This article is included in the Quantum Technology collection.
As graph datasets—including social networks, supply chains, and bioinformatics data—grow in size and complexity, researchers are driven to search for solutions enhancing model efficiency and speed. One avenue that may provide a solution is Quantum Graph Learning (QGL), a subfield of Quantum Machine Learning (QML) that applies machine learning inspired or powered by quantum computing to graph learning tasks.
We reevaluate Quantum Feature Embeddings (QFE), a QGL methodology published by Xu et al. earlier this year. QFE uses Variational Quantum Circuits to preprocess node features and then sends them to a classical Graph Neural Network (GNN), with the goal of increasing performance and/or decreasing total model size. Xu et al. evaluated this methodology by comparing its performance with the performance of variously-sized classical models on the benchmark datasets PROTEINS and ENZYMES, and they report success.
Our core methodology and learning task remain unchanged. However, we have made several changes to the experimental design that enhance the rigor of the study: 1) we include the testing of models with no embedder; 2) we conduct a thorough hyperparameter search using a state-of-the-art optimization algorithm; and 3) we conduct stratified five-fold cross-validation, which mitigates the bias produced by our small datasets and provides multiple test statistics from which we can calculate a confidence interval.
We produce classical models that perform comparably to QFE and significantly outperform the small classical models used in Xu et al.’s comparison. Notably, many of our classical models achieve this using fewer parameters than the QFE models we trained. Xu et al. do not report their total model sizes.
Our study sheds doubt on the efficacy of QFE by demonstrating that small, well-tuned classical models can perform just as well as QFE, highlighting the importance of hyperparameter tuning and rigorous experimental design.
Quantum Machine Learning, Quantum Graph Learning, Graph Neural Networks, Quantum Feature Embedding
Over the last two decades, a new field at the intersection of quantum computing and machine learning has emerged: Quantum Machine Learning (QML). The methods in this field are best understood when broken down into three categories, as described by Houssein et al.1:
1. Pure QML: algorithms that rely solely on quantum circuits, like the quantum version of the support vector machine,
2. Quantum-inspired ML: classical machine learning that takes inspiration from the field of quantum computing,
3. Quantum-Classical Hybrid ML: techniques that use both quantum computing and classical computing; this includes Variational Quantum Circuits (VQCs) — quantum circuits with classical parameters that can be updated during training using a classical optimizer.
Algorithms in each of these categories can be applied to a variety of tasks including classification, regression, and optimization.1
Within QML, an even newer subfield called Quantum Graph Learning (QGL) is starting to be explored. As described by Yu et al.,2 QGL has the potential to solve or mitigate several substantial problems in graph learning including the difficulty of storing and processing large graphs and the limitation of the distance across which inferences can be made. QGL should also be able to apply some of the native benefits of QML to graph learning, including a reduction in the number of required training parameters.
Yu et al.2 describe several QML techniques that can be applied to graph learning, including various types of quantum optimization, quantum random walks and quantum graph kernels (used to represent graphs in the vector space of quantum circuits), and VQCs.
The Quantum Feature Embeddings (QFE) methodology proposed by Xu et al.3 is a special case of the VQC technique. It applies a VQC to the node features of the graph input to generate embeddings, which are then passed to a classical message-passing model (e.g. a Graph Convolutional Network4). This is depicted in Figure 1. (Note that an additional pooling layer has been included. Although this was not mentioned by Xu et al.,3 it is necessary for graph classification.)
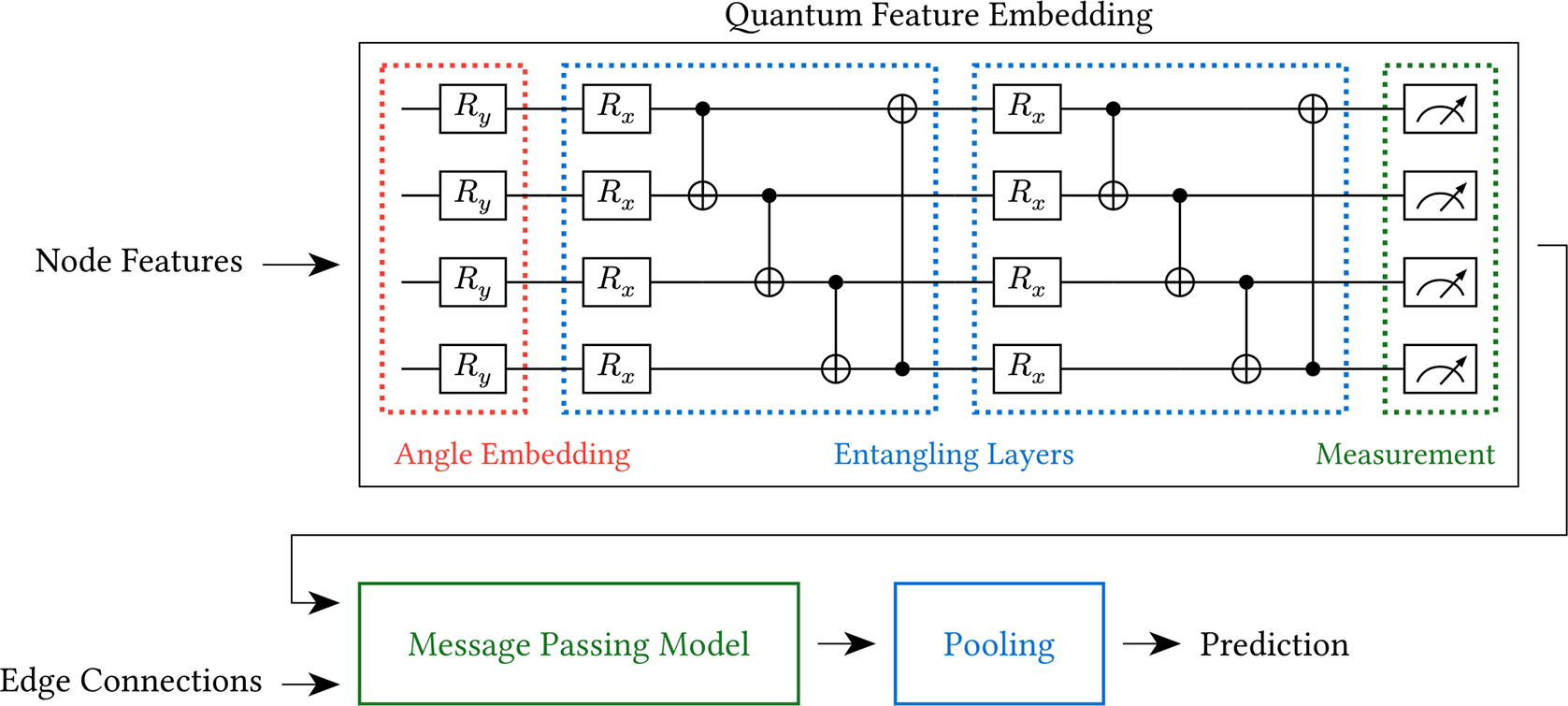
Xu et al. argue that using a VQC to create embeddings provides several benefits. First, because of the unitary nature of the circuit, the norms of the embeddings are preserved. This preservation helps increase the stability of the model during training. The unitary nature of the VQC also implies that it acts as a bijective function, which means that distinct inputs will not be mapped to the same output. This implies that information will not be lost during the embedding process. Finally, the exponential nature of the circuit’s vector space allows the VQC to approximate complex functions with exponentially fewer trainable parameters compared to classical models.
To test QFE empirically, Xu et al. evaluated its performance on the classification problems defined by two benchmark datasets: PROTEINS5,6 and ENZYMES.5,7 That performance was then compared with the performance of two classical embedders using Multi-Layer Perceptrons (MLPs). One had hidden nodes (to approximate the number of parameters in QFE), where is the number of input features; the other had hidden nodes (to approximate the expressive power of QFE’s vector space). Notably, Xu et al. did not compare QFE with a model with no embedding layer.
Their results, as depicted in Figure 2, led them to conclude that QFE provides an increased accuracy compared to classical models with similar numbers of parameters and can keep up with classical models that have exponentially more parameters.
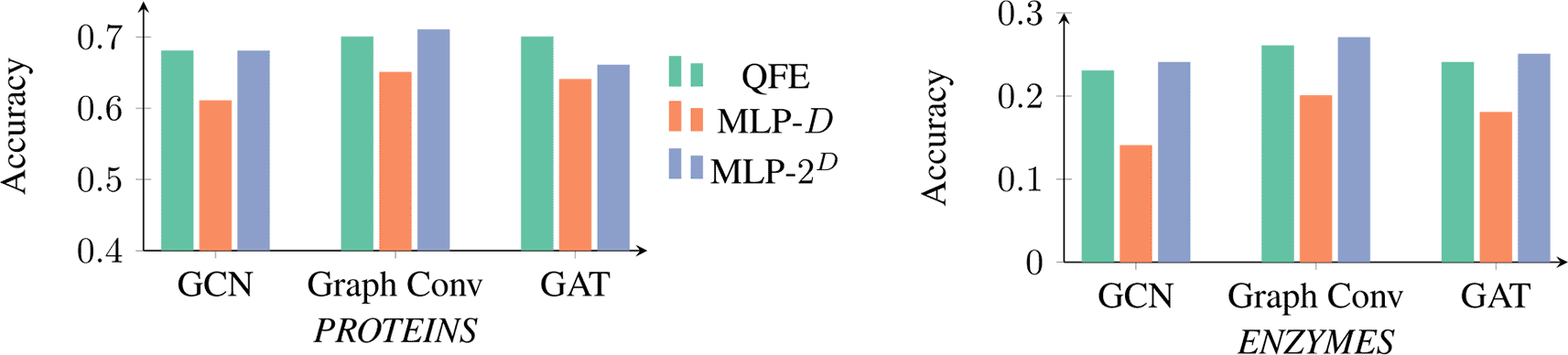
Note: The State-Of-The-Art (SOTA) performance on PROTEINS is 8 which is much higher than what QFE achieved. However, because QGL and QML in general are young fields, we do not necessarily expect QML models to achieve SOTA performance. Instead, we simply want to determine if the methodology has a positive impact and if it might be useful in the future.
We critically examine QFE using an improved experimental design with state-of-the-art hyperparameter tuning. This approach yields classical models that perform comparably to QFE and significantly outperform the small classical models used in Xu et al.’s comparison. In addition, we demonstrate that it is possible to obtain similar accuracies with classical models that have fewer trainable parameters.
While this paper may not explore new and exciting QML methodologies, it does provide a rigorous evaluation of an existing technique, which is scientifically valuable. Without negative and critical results like this one, it would be impossible to allocate precious research resources efficiently.
The structure of the individual models we tested remains largely unchanged from the methodology created by Xu et al. During inference, node features are taken from the input graph (or batch of graphs) and passed to the embedder component (if it exists). There are five options for the embedder component:
1. No embedder component,
2. MLP-, a 2-layer perceptron with input nodes, hidden nodes and output nodes,
3. MLP-, a 2-layer perceptron with input nodes, hidden nodes, and output nodes,
4. QFE-exp, which measures the expectation value of each wire with respect to the Pauli Z gate, and
5. QFE-probs, which measures the probability of each possible output (measured in the computational basis).
The QFE embedder, a VQC, starts with an angle embedding component using gates (defined below). This component is used to convert the classical node features into a state the quantum computer can process.
In the entangling component, parameterized gates and controlled not gates (defined below) are combined to create entangling layers which allow the circuit to represent complex functions. The parameters (i.e. the many instances of ) in these layers are used to update the circuit based on loss calculated between the circuit’s output and the expected output.
The number of layers used in the entangling component is a hyperparameter of our model. As in Xu et al.’s article,3 we finish with a measuring component that measures all of the wires in the quantum circuit.
Following that, the embeddings and the edge connections from the input graph are given to the message-passing model. This model can use any type of message-passing layer (including any one of the convolutional layers provided by the PyTorch Geometric python package9). We chose to test the three layer types chosen by Xu et al.3:
The number of hidden channels and the number of layers used by the message-passing model are hyperparameters. The number of output channels is equal to the number of classes in the classification problem.
Next, a pooling layer is used to aggregate the data spread across the nodes of the graph being processed. We tested the three simple options below.
Finally, a log Softmax function is applied.
During initial training runs, we tried both cross-entropy loss, which was used by Xu et al.,3 and Negative Log Likelihood (NLL) loss, which was used in PyTorch Geometric9 examples and in HGP-SL,8 the classical model which achieved the SOTA accuracy on the PROTEINS dataset. Both losses produced similar results; we chose to use NLL loss for its popularity.
To optimize the parameters of the entire model (including the QFE embedder, when applicable), we used Adam.12
To mitigate overfitting, we implemented early stopping with a patience of epochs and a maximum training duration of epochs. The metric used for early stopping is the loss taken from the validation dataset. We also employed dropout, the rate of which is a hyperparameter of the model.
For each embedder option, we optimized our hyperparameters by running a multi-objective Optuna13 study that used the Non-dominated Sorting Genetic Algorithm II (NSGA-II) algorithm.14 Each study included approximately experiments. Their objectives were to …
• maximize average validation accuracy,
• minimize the variability of the validation accuracy (based on a confidence interval),
• minimize the number of trainable parameters.
The hyperparameters we optimized are listed below.
• The number of QFE layers (if applicable)
• The message passing layer type
• The number of layers and hidden channels in the message-passing model
• The dropout rate
• The Adam optimizer’s learning rate and weight decay
We also optimized the batch size during our initial experiments. However, since this hyperparameter was shown to have low importance (using the fANOVA evaluation method15) and since the varying memory usage it caused made it difficult to run experiments in parallel, we ultimately chose to hold it constant at .
To increase the stability of each experiment within the Optuna13 study and obtain an estimate of variability, we performed stratifiedfive-fold cross-validation on the provided training dataset and then bootstrapped the accuracies to obtain a confidence interval.
Stratified five-fold cross-validation was conducted on the entire dataset to control for the bias caused by our train/test data split and to provide multiple test accuracies which could be used to estimate the variability of the test statistic.
For each of the train/test data splits provided by cross-validation and for each embedder option, we ran hyperparameter tuning and then selected three of the best models produced, each maximizing one of the utility functions below.
where is the z-score of the model’s validation accuracy, is the z-score of the model’s accuracy variability, and is the z-score of the model’s trainable parameter count. All z-scores [1] are taken with respect to the distribution of experiments at the Pareto front of the corresponding Optuna13 study.To decrease the storage and maintenance burden during hyperparameter tuning and because each model only took a few minutes to train [2], we chose not to save model weights. Instead, we retrained each of the selected models after tuning, this time using a stratified shuffle split to divide the training dataset into a training dataset and a validation dataset (for use during early stopping). To decrease the non-deterministic effects of our training script on the results, we also implemented a minimum training duration of 60 epochs. If early stopping was triggered before then (which likely indicates that training had failed to produce a well-performing model), then the model would be retrained. This could happen up to five times before the training script progresses to the next model.
Finally, we evaluated the selected models from each fold on their corresponding test dataset and then used bootstrapping to calculate a confidence interval for the mean test accuracy.
To evaluate our models, we used the same two protein-related graph datasets used by Xu et al.3: PROTEINS5,6 and ENZYMES.5,7 Both of these datasets are a part of the TUDataset collection16 and are publicly available on http://www.graphlearning.io.
The graphs contained in these datasets represent the structures of proteins, and the nodes within these graphs represent secondary protein structures — helixes, sheets, and turns. Two nodes are connected “if they are neighbors along the amino acid sequence or one of three nearest neighbors in space”.16 In addition to the secondary structure type, which is represented in the dataset using one-hot encoding, nodes have feature(s) representing some of the structure’s physical and/or chemical properties. The PROTEINS dataset has one such feature; the ENZYMES dataset has 18. Because our computer could not simulate the 21 qubits (three one-hot encoded categories + 18 additional features) that would be required for the ENZYMES dataset, we used Principal Component Analysis (PCA) to reduce the node features back down to four dimensions. Despite this large reduction, the resulting features explain of the variance in the original features.
In PROTEINS, proteins are classified as enzymatic or non-enzymatic. In ENZYMES, proteins (which are all enzymatic) are classified according to the class of the reaction they catalyze; there are six such classes. For both of these datasets, our goal is to predict the graph-level classifications based on the nodes’ features and graph’s structure as a whole.
Both datasets are quite small; PROTEINS contains graphs and ENZYMES contains . And the graphs in both datasets are mid-sized; in PROTEINS, they have an average of nodes and edges, and in ENZYMES, the have an average of nodes and edges. A few examples from PROTEINS and ENZYMES are depicted in Figures 3 and 4 respectively.
This section focuses on notable examples from and visual representations of our results. Our complete results data, optimized hyperparameters, model weights, and code have been published on Zenodo17 under the Creative Commons Attribution 4.0 International Public License (CC BY 4.0). Our code is also available on GitHub at https://github.com/jsimonrichard/QFE-Experiments under the MIT License.
Using , we found models with average test accuracies on the PROTEINS dataset ranging from to . In particular, we achieved an average accuracy of using the QFE-probs embedder and an average accuracy of using the QFE-exp embedder, both of which are comparable to the results achieved by Xu et al.3 However, we also achieved an average accuracy of using the MLP- embedder and an average accuracy of using no embedder, which are comparable to the QFE accuracies and significantly higher than the MLP- accuracies produced by Xu et al.3
The average sizes of our models range from 111K to 320K trainable parameters. Notably, the average number of trainable parameters used by our MLP--based models is lower than the average for our QFE-based-models (but not significantly lower) even though it exceeds their average accuracies. The results for all five embedder types are depicted with error bars () in Figure 5.
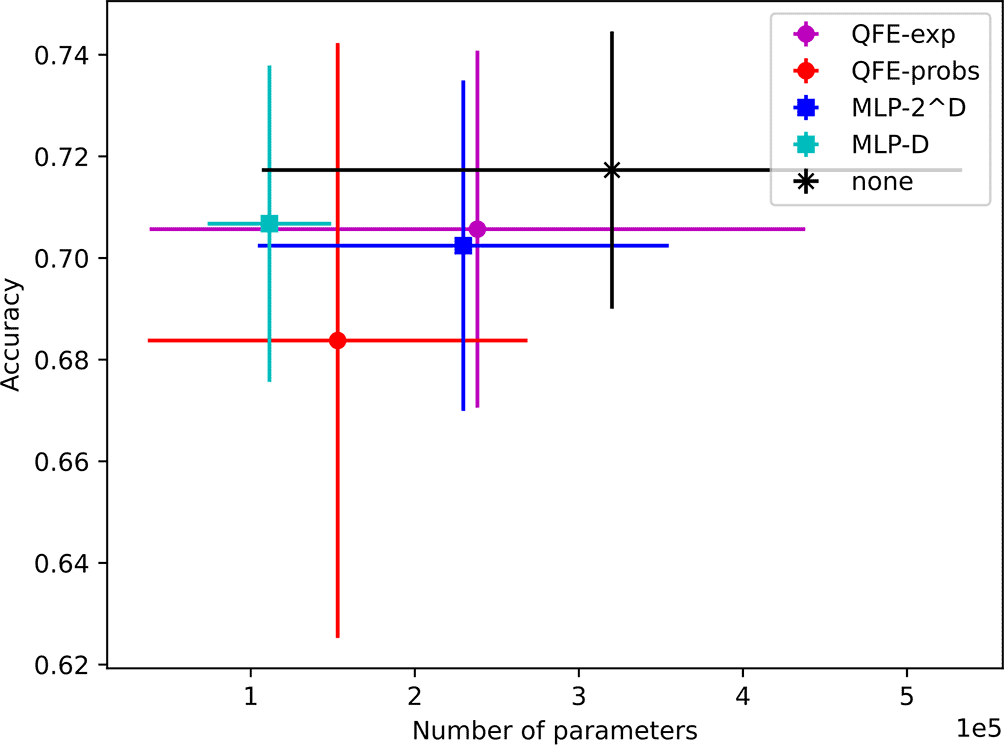
Unfortunately, Xu et al. did not report their models’ total sizes, so we cannot make direct size comparisons between our models and theirs. They did report the number of floating-point operations (FLOPS) required for MLP- embedder to process a graph with nodes and the quantum gate operations required for their QFE embedder to process a graph of the same size.3,18 However, these metrics are not relevant to our analysis since the sizes of the embedders are overshadowed by the variance in the sizes of the message-passing layers.
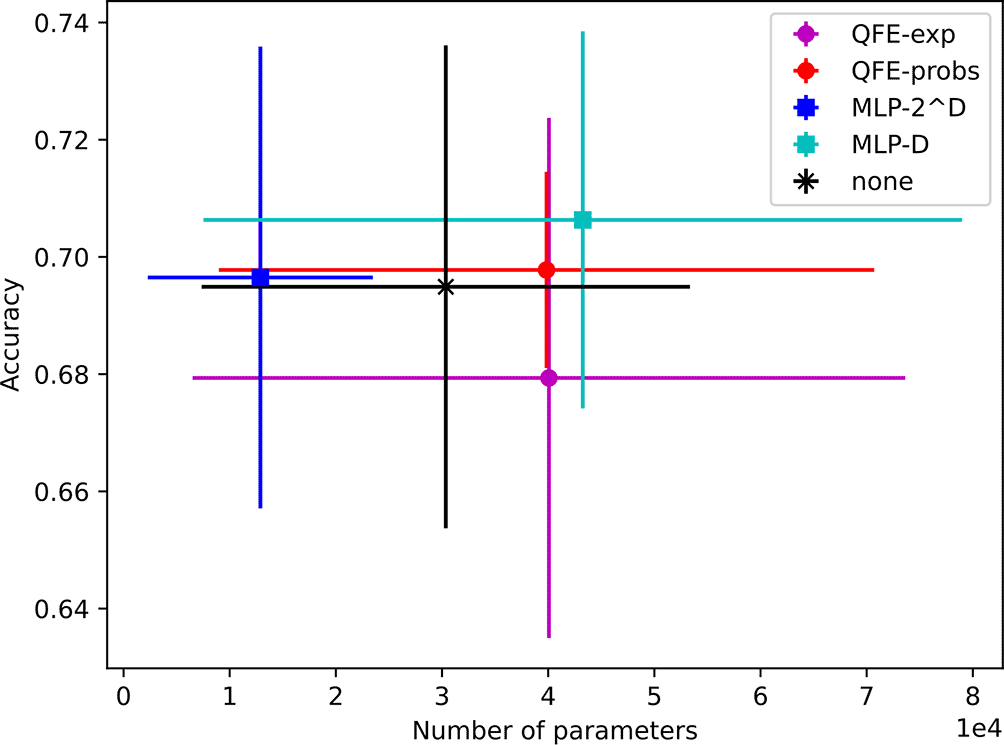
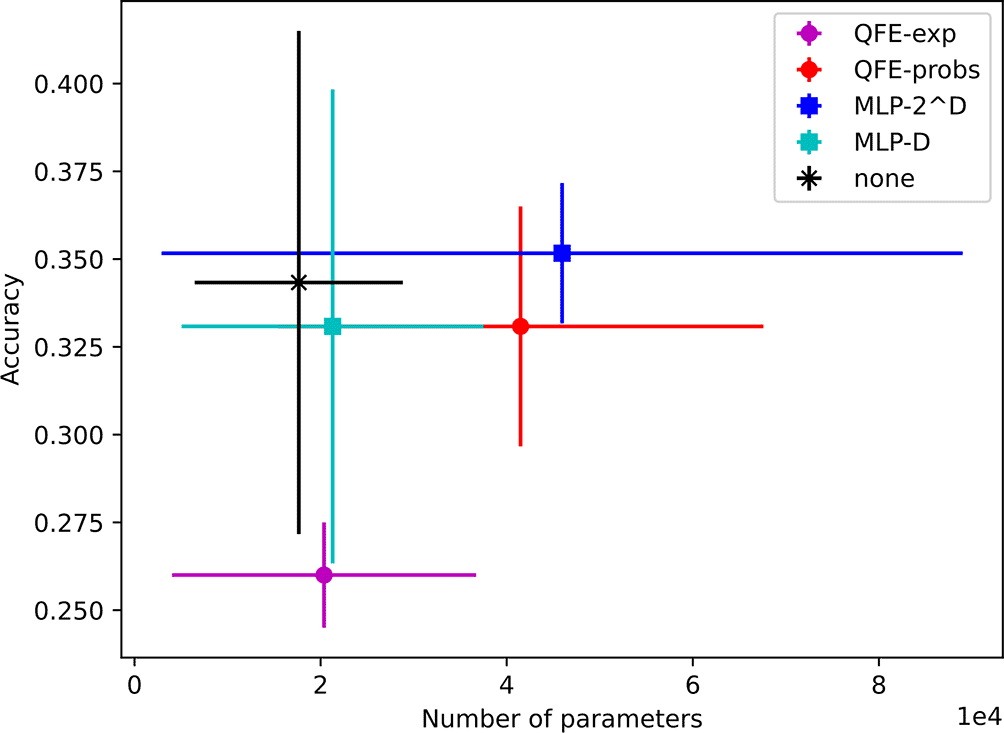
The models found using performed almost as well as those found with , but with an order of magnitude fewer parameters. They achieve accuracies ranging from to with average parameter counts ranging from 13K to 43K. Their results are depicted in Figure 6.
Finally, we found models using that maintain that performance with even fewer parameters. Their average accuracies range from to and their average sizes range from 3K to 11K trainable parameters. Notably, this includes the models using MLP-, which maintain an average accuracy of using an average of 3K trainable parameters. The results for all five embedder types are shown in Figure 7.
Our results from ENZYMES represent an even more drastic departure from the results reported by Xu et al. In fact, every single average taken from the models using MLP- or no embedder is significantly greater than the highest MLP- accuracy that Xu et al. achieved [3].
The models found using achieve average accuracies ranging from to with average parameter counts ranging from 124K to 299K. Notably, the models with no embedder achieved the highest average accuracy () with an average of 269K trainable parameters. This average is significantly greater than all of the accuracies reported by Xu et al. The results for all five embedders are depicted in Figure 8.
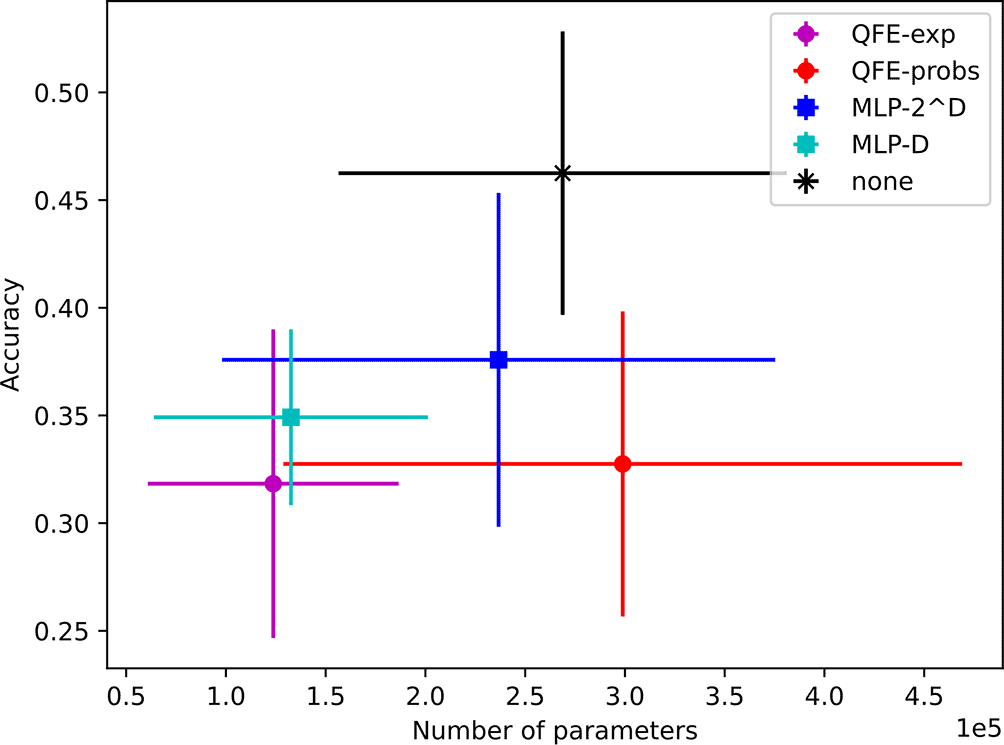
Again, most of the models found using achieve similar average accuracies (although the highest average is no longer ) using an order of magnitude fewer parameters. Their average accuracies range from to and their average parameter counts range from 18K to 46K. These results are shown in Figure 9.
Finally, the models found by achieve similar results with marginally fewer parameters for some and essentially the same number of parameters for others. They achieve accuracies ranging from to with sizes ranging from 3K to 48K trainable parameters. These results are shown in Figure 10.
Our analysis sheds doubt on QFE’s efficacy in the real world despite its interesting mathematical foundations. Using the same datasets as Xu et al.,3 we achieved similar and, in some cases, significantly better performance compared to the accuracies reported by Xu et al.3 using models with QFE, models with the MLP- embedder, and models with no embedder at all. In addition, we showed that it is possible to find classical models with comparable performance that have fewer trainable parameters than their quantum-classical hybrid counterparts. Why did Xu et al. fail to reach these conclusions? There may have been a few contributing factors.
• They overlooked classical models with no external embedder.
• They probably did not use the same hyperparameter tuning method we did (they make no mention of their tuning methods in3).
In addition, they did not provide error bars, so it is hard to know whether their results are statistically significant.
Our study highlights the critical importance of careful experimental design, statistical testing, and thorough hyperparameter tuning. Without these things, it is difficult if not impossible to draw sound conclusions in a rapidly evolving field like QGL.
There are several limitations (and corresponding future research directions) to this study.
• Because of the small sizes of both the PROTEINS and ENZYMES datasets, it is difficult to show statistically significant differences. We were able to do so in a few cases, but most comparisons that could be made are insignificant. Future research may include evaluating QFE on larger datasets.
• It is still unclear why QFE is failing to provide an advantage in either accuracy or model size. One possibility is that the QFE embedder does not have much information to work with because it only operates on individual node features, which are fairly one-dimensional (in both PROTEINS and ENZYMES, the first component generated by PCA explains over of the variance). QFE may still be useful in other contexts.
Our code can be found on GitHub: https://github.com/jsimonrichard/QFE-Experiments. The main libraries we used are listed below; thanks go out to all of their authors and maintainers.
In addition, our training script was largely inspired by the training script written for HGP-SL16 (located at https://github.com/cszhangzhen/HGP-SL).
Our complete results data, optimized hyperparameters, model weights, and code have been published on Zenodo with the title “Trained models, code, result data, and Optuna study data from ‘Hybrid quantum or purely classical? Assessing the utility of quantum feature embeddings.’” 17 doi: 10.5281/zenodo.13117645 under the Creative Commons Attribution 4.0 International Public License (CC BY 4.0). Our code is also available on GitHub at https://github.com/jsimonrichard/QFE-Experiments under the MIT License.
1. ENZYMES: http://www.graphlearning.io/ ; publicly available, but attribution is required (see https://chrsmrrs.github.io/datasets/docs/home/ )
2. PROTEINS: http://www.graphlearning.io/ ; publicly available, but attribution is required (see https://chrsmrrs.github.io/datasets/docs/home/ )
This paper is the primary product of J. Simon Richard’s undergraduate Honors Thesis for the Washkewicz College of Engineering at Cleveland State University. Thank you to the Washkewicz College of Engineering for its support.
In addition, thank you to Sascha Xu, a current PhD student at the CISPA Helmholtz Center for Information Security and the corresponding author of Xu et al.’s article, 3 for his quick email responses and for his willingness to help.
Finally, we acknowledge that AI—specifically Anthropic’s Claude 3.5 Sonnet model—was used to help draft and/or improve the phrasing of a few sections in this paper: the abstract, section 1.2, and the last paragraph of section 5. Each of those sections have been reviewed and edited thoroughly for clarity and correctness by the human author of this paper.
1 We chose to use z-scores because they are simple, scale-independent, and less affected by outliers compared to min-max normalization. However, it is important to note that the underlying distributions are not normal, so we cannot use these scores to analyze the experiment distributions directly.
2 On our machine—a Linux PC with an AMD Ryzen Threadripper 1920X CPU and an Nvidia RTX 2080 Ti GPU—the quantum-classical hybrid models took an average of 8 minutes to train on PROTEINS and an average of 3 minutes and 18 seconds to train on ENZYMES. The purely classical models took an average of 1 minute and 14 seconds to train on PROTEINS and an average of 1 minute and 25 seconds to train on ENZYMES.
3 Xu et al. did not report the raw data for their ENZYMES results, so we took them from their results plot (Figure 2) using https://plotdigitizer.com/app. The values we measured can be found in our Zenodo repository. 17
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Yang C.-H. H, Qi J, Chen S, Chen PY, et al.: Decentralizing Feature Extraction with Quantum Convolutional Neural Network for Automatic Speech Recognition. IEEE. 2021. 6523-6527 Reference SourceCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: quantum machine learning, machine learning, quantum computing
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Ma Y, Tresp V: Quantum Machine Learning Algorithm for Knowledge Graphs. ACM Transactions on Quantum Computing. 2021; 2 (3): 1-28 Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Quantum Machine Learning, Temporal and dynamic graphs
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 23 Aug 24 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)