Keywords
Alopecia, cynomolgus macaques, fungi, skin mycobiota, Oxford Nanopore Technologies
This article is included in the Pathogens gateway.
This article is included in the Nanopore Analysis gateway.
Alopecia in captive non-human primates (NHPs) is a major animal welfare concern in many primate facilities. Some studies suggested that the mycobiome, considered fungal communities in specific areas, could influence the pathogenesis of alopecia. However, the pathogenesis mechanism of alopecia in cynomolgus macaques is poorly known.
This study aims to investigate the association between alopecia and skin mycobiota in cynomolgus macaques.
Skin swab samples were collected from five areas where alopecia is common (arm, dorsal, head, leg, and ventral areas) of 47 healthy and 50 alopecic macaques. The samples were extracted for DNA. Oxford Nanopore Technologies (ONT) was applied to explore the skin mycobiota based on full-length Internal Transcribed Spacer (ITS) sequencing.
Our findings showed that alpha and beta diversities significantly differed between alopecic and healthy macaques among the five areas. Wallemia sebi was associated with alopecia in the arm. Furthermore, Candida albicans, Rhodotorula mucilaginosa, Exophiala dermatitidis, and Candida parapsilosis were prevalent in the dorsal and head areas. Moreover, Aspergillus penicillioides was dominant in the dorsal and ventral areas. Pseudozyma sp., Moesziomyces antarcticus, and Cladosporium dominicanum were uniquely found in the head area. Apiotrichum domesticum was highly observed in the leg and ventral areas. Lastly, Cladosporium halotolerans was uniquely detected in the ventral area.
These microorganisms may be associated with the development of alopecia in cynomolgus macaques. These findings might be useful for biomedical research and therapeutic management strategies of animal health in primate facilities in the future.
Alopecia, cynomolgus macaques, fungi, skin mycobiota, Oxford Nanopore Technologies
Alopecia (hair loss) is a dermatological disorder that is a frequent occurrence in some non-human primates (NHPs) housed in captivity,1 which the genus Macaca is an essential NHPs model and is widely used in biomedical research, human health, disease mechanisms, particularly for human pharmaceutical testing2 such as cynomolgus macaques (M. fascicularis) and rhesus macaques (M. mulatta). Moreover, veterinarians often report alopecia in these species. Previous studies indicated that 34–86.5% of laboratory-housed rhesus macaques exhibit some degree of alopecia.3 Lesional alopecia can be developed on the skin at any stage of the lifespan.4 Furthermore, the pattern of hair loss can range from small patches to extensive areas covering a significant portion of the animal’s body, potentially reducing the overall quality of life.3 Alopecia impacts not only the health of the animals but also the reliability of experimental results.5 Therefore, this condition raises significant animal welfare concerns in many primate facilities. Alopecia can be caused by a variety of factors, such as demographics, skin disorders, immunological conditions, direct skin contact with allergens, inflammation, physiological issues, environmental factors, and grooming or hair-plucking behaviors.1,4–7 For instance, rhesus macaques’ dorsal hair loss in research facilities was caused by hair-pulling or over-grooming by cage mates.8 Some studies suggested that the mycobiome, considered fungal communities in specific areas, could influence the pathogenesis of alopecia.4,9 However, the pathogenesis mechanism of alopecia in cynomolgus macaques is poorly known. Consequently, it is crucial to investigate the potential causes of alopecia and develop effective treatments for affected animals.
In a particular environment, the microbiome encompasses all communities of commensal, symbiotic, and pathogenic microbes, including fungi.10 All layers of the skin are typically associated with the microbiome and produce ecological conditions that support fungal colonization. Consequently, disruption or imbalance between unhealthy and healthy microbes in the skin may contribute to the development of alopecia. Previous reports in rhesus macaques showed that an overgrowth of fungi on the skin, including Microsporum canis and Trichophyton mentagrophytes, may also lead to alopecia. Additionally, Candida albicans, an opportunistic pathogen, is frequently detected in immunocompromised and debilitated individuals, resulting in superficial infections of the skin and mucous membranes. Opportunistic fungal skin infections, which may be associated with fighting wounds or trauma, combined with environmental factors and immunological disorders, are thought to contribute to the pathogenesis of alopecia.9 Although fungal infections may contribute to the pathogenesis of alopecia, no previous published reports have identified them as a leading cause of alopecia in cynomolgus macaques. Therefore, these factors need to be examined in more detail, as exploring the potential association between fungal infections and hair loss in alopecic macaques is of significant interest.
The accurate identification of pathogenic diseases is essential for the effective treatment and diagnosis of infections. Currently, numerous approaches are available to characterize the composition of microorganisms and identify potential pathogens, such as high-throughput sequencing (HTS), polymerase chain reaction (PCR), and microbial culture.11 Microbial culture is regarded as the gold standard among diagnostic methods. However, culture techniques may not successfully detect unculturable, unknown, or novel pathogens.12 PCR assays offer a low-cost, rapid, and specific method for amplifying target nucleic acid sequences. On the other hand, the throughput capabilities of PCR are relatively limited.13,14 HTS, also known as next-generation sequencing (NGS), is a novel technique for DNA and RNA sequencing, as well as variant and mutation detection. There are various HTS technologies, such as Illumina and Oxford Nanopore Technologies (ONT), that can rapidly sequence hundreds to thousands of genes or whole genomes within a short period and are cost-effective.15 Illumina is a second-generation approach to short-read sequencing that provides high accuracy. However, this technique remains limited due to short read lengths and lacks the taxonomic resolution required for precise species-level identification in closely related phylogenetic groups.16,17 ONT is a third-generation long-read sequencing that has provided higher efficiency data output. However, the primary limitation of ONT was a high error rate. Therefore, the error rate is improved by improving the chemistry used in bioinformatic pipelines.17 ONT is capable of full-length sequencing of the fungal ITS region (~800 bp, covering ITS1 and ITS2).18 Moreover, ONT sequencing demonstrated a greater resolution in fungal identification at the species level than short-read sequencing, which is limited to read lengths of approximately 300–500 bp.19 This enables precise identification of fungi for mycobiome studies.
Therefore, this study aims to compare mycobiota composition, abundance, and profile between alopecia and healthy skin cynomolgus macaques, utilizing ONT sequencing to analyze the fungal ITS region. This finding could potentially be used to inform future treatments for alopecia. It could help to discover a new model for understanding alopecia pathogenesis and develop novel therapeutics for skin diseases that are linked to mycobiome dysbiosis in macaques.
The animal subjects enrolled in this study were cynomolgus macaques (M. fascicularis) housed at NPRCT-CU, Saraburi, Thailand (GPS: 14° 52’ N, 100° 90’ E). The housing system of macaques was strictly hygienic and conventional at the NPRCT-CU. Macaques were housed in stainless-steel social cages (4 × 4 × 3–3.5 m; width × length × height) containing 3–15 animals per cage. The housing environment was maintained at 21–39°C with 40–70% humidity under standard fluorescent lighting. Animals were fed standard monkey chow (Perfect Companion Group Co., Ltd) in the morning and fresh fruits from local suppliers in the afternoon, with hyperchlorinated water (1 ppm; pH 7.3–7.7). The study protocol relating to macaques was approved by the Animal Care and Use Committee of the National Primate Research Center of Thailand Chulalongkorn University (Protocol Review No. 2375015). All animal experiments were performed following the ARRIVE guidelines (https://arriveguidelines.org/arrive-guidelines ). The sample size was determined using Cochran’s formula,20 a method commonly applied when the population size is large or unknown. For anesthesia, the macaques were caught from the gang cage using a scoop net. They were intramuscularly injected with 2-5 mg/kg of tiletamine–zolazepam (ZoletilTM) mixed with 0.02-0.05 mg/kg of Dexmedetomidine during a semi-annual health check. After all sample collection had been completed together with other routine procedures in NPRCT-CU, the macaques were moved back to the gang cage area and injected with Atipamezole (equal volume (ml) to Dexmedetomidine) to reverse the effects of Dexmedetomidine. These procedures were carried out in accordance with the NPRCT-CU, which is accredited by AAALAC and adheres to OECD-GLP international standards. The animals were categorized into two age classes: Juvenile/Subadult (6 years or younger) and Adult (older than 6 years). Exclusion criteria for both male and female macaques were tuberculosis, SRV, SIV, and STLV, and pregnancy or lactation for females. The details of the body part with alopecia and the demographics of macaques are shown in Table 1 and Supplementary Table 1.
Skin swabs were collected from 97 cynomolgus macaques during annual health checks of animals. Animals were divided into two groups: healthy (n = 47) and alopecic animals (n = 50). As alopecia mainly occurred on the arm, dorsal, head, leg, and ventral areas, each healthy, non-alopecic control monkey was swabbed in those 5 areas. Alopecic animals were collected skin swab samples only at the lesion area, varied from 1 to 5 areas per macaque ( Table 1). Before sample collection, the cotton swabs (Puritan™ Medical Product, USA) were soaked with phosphate-buffered saline (PBS). All samples were stored with 1 ml of Nucleic Acid Preservation (NAP) buffer (Cat. No. 1-NAP-100, BioEntist, Thailand) in 2 ml conical tubes at -20°C until used for DNA extraction and long-term storage.
The DNA was extracted using the ZymoBIOMICSTM DNA Miniprep Kit (Cat. No. D4300, Zymo Research, USA) according to the manufacturer’s protocol. The extracted DNA was stored at -20°C until used. The ITS region (ITS1 and ITS2) of fungal nuclear DNA was amplified using primers modified from a previous study.21,22 These primers included 3’ specific target sequences (underlined) and 5’ nanopore adaptors, as follows: For fungi, ITS_1F: 5’-TTTCTGTTGGTGCTGATATTGCTCCG TAGGTGAACCTGCGG-3’ and ITS_4R: 5’-ACTTGCCTGTCGCTCTATCTTC TCCTCCGCTTATT GATATGC-3’. The 10 μl PCR reaction contains 5 μl of KOD One PCR Master Mix-Blue (Cat. No. KMM-201, TOYOBO, Japanese), 0.3 μl each of forward and reverse primer, 3.4 μl of ddH2O, and 1 μl of DNA template. The thermal condition for PCR consists of 1 cycle of initial denaturation at 98°C for 2 min, followed by 35 cycles of 98°C for 10 s, 60°C for 10 s, 68°C for 10 s, and 1 cycle of final extension at 68°C for 5 min, respectively. Then, the PCR products were amplified with the multiplexing barcodes using the PCR Barcoding Expansion 1-96 kit (Cat. No. EXP-PBC096, Oxford Nanopore Technologies, UK). The PCR condition of the barcoding step was initial denaturation at 98°C for 2 min, then 5 cycles of 98°C for 10 s, 60°C for 10 s, followed by 68°C for 10 s, and final extension at 68°C for 5 min, respectively. The barcoded PCR products were analyzed using 1% agarose gel electrophoresis. The DNA library was purified using the QIAquick PCR Purification Kit (Cat. No. 28106, Qiagen, Germany) according to the manufacturer’s protocol. The quantity of DNA was measured using the QubitTM dsDNA HS Assay Kit (Cat. No. Q32854, Invitrogen, USA) with the Qubit 4.0 Fluorometer (Invitrogen, USA). Then, products were equimolarly pooled and size-selected using 0.8x Agencourt AMPure XP beads (Cat. No. A63882, Beckman Coulter, USA). The pooled library was ligated with Nanopore adaptors using the SQK-LSK114 Ligation Sequencing Kit (Cat. No. SQK-LSK114, Oxford Nanopore Technologies, UK) for nanopore sequencing according to the manufacturer, and the sequencing was performed on the MinION Mk1C sequencer (Oxford Nanopore Technologies, UK) using the FLO-MIN114 flow cell (Oxford Nanopore Technologies, UK).
Basecalling of the FAST5 files was carried out using Guppy basecaller software v6.5.723 (Oxford Nanopore Technologies, UK) to generate FASTQ sequences with super-accuracy (SUP) mode, applying a minimum quality threshold of Q>15. The quality of reads was analyzed by MinIONQC v1.4.1.24 Then, the FASTQ sequences were demultiplexed by Porechop v0.2.4 (https://github.com/rrwick/Porechop). Clustering, polishing, and taxonomic classification were performed using the NanoCLUST pipeline.25 ITS regions of fungal rDNA sequences were identified using the SILVA database.26 The fungal abundance data were subsequently examined using QIIME2 software v2021.227 to generate a collapsed taxa table. The rarefaction curve, relative abundances, linear discriminant analysis effect size (LEfSe), and alpha diversity with Chao and Shannon indexes were visualized by MicrobiomeAnalyst.28 The Mann-Whitney U test was used for the statistical analysis of alpha diversity between skin mycobiota and macaque groups. Beta diversity was analyzed using Bray-Curtis dissimilarity, with statistical comparisons of mycobiome profiles performed using PERMANOVA (P < 0.05). Differential enrichment of fungal communities was analyzed using LEfSe with the threshold of Linear discriminant analysis (LDA) score >3 and P < 0.05 (Kruskal-Wallis test).
The rarefaction curve demonstrated a gradual flattening, which indicated that the sequencing data had reached saturation, covering most fungi species presented in different body parts of both healthy and alopecic cynomolgus macaques, as shown in Supplementary Figure 1. The rarefaction curve for the alopecic group is lower than that of the healthy group, indicating that the alopecic group has less diversity of fungi.
Alpha diversity comparisons across five body parts between healthy and alopecic macaques revealed statistically significant differences in the Chao1 and Shannon indexes (Kruskal-Wallis test) in Figure 1a and 1b. The Chao1 diversity index was significantly higher in the dorsal (P = 4.65 x 10−5), head (P = 5.53 x 10−4), and ventral (P = 4.39 x 10−5) areas of the alopecic group than in the healthy group. However, the Shannon diversity index of the alopecic group was significantly lower than that of the healthy group in the arm area (P = 8.45 x 10−3). Notably, the Shannon diversity index in the dorsal area of the alopecic group was significantly higher than the healthy group (P = 7.16 x 10−4). Beta diversity was significantly different between healthy and alopecic cynomolgus macaques in the arm (P = 0.001), dorsal (P = 0.001), head (P = 0.001), leg (P = 0.001), and ventral area (P = 0.017) in Figure 1c-1g.
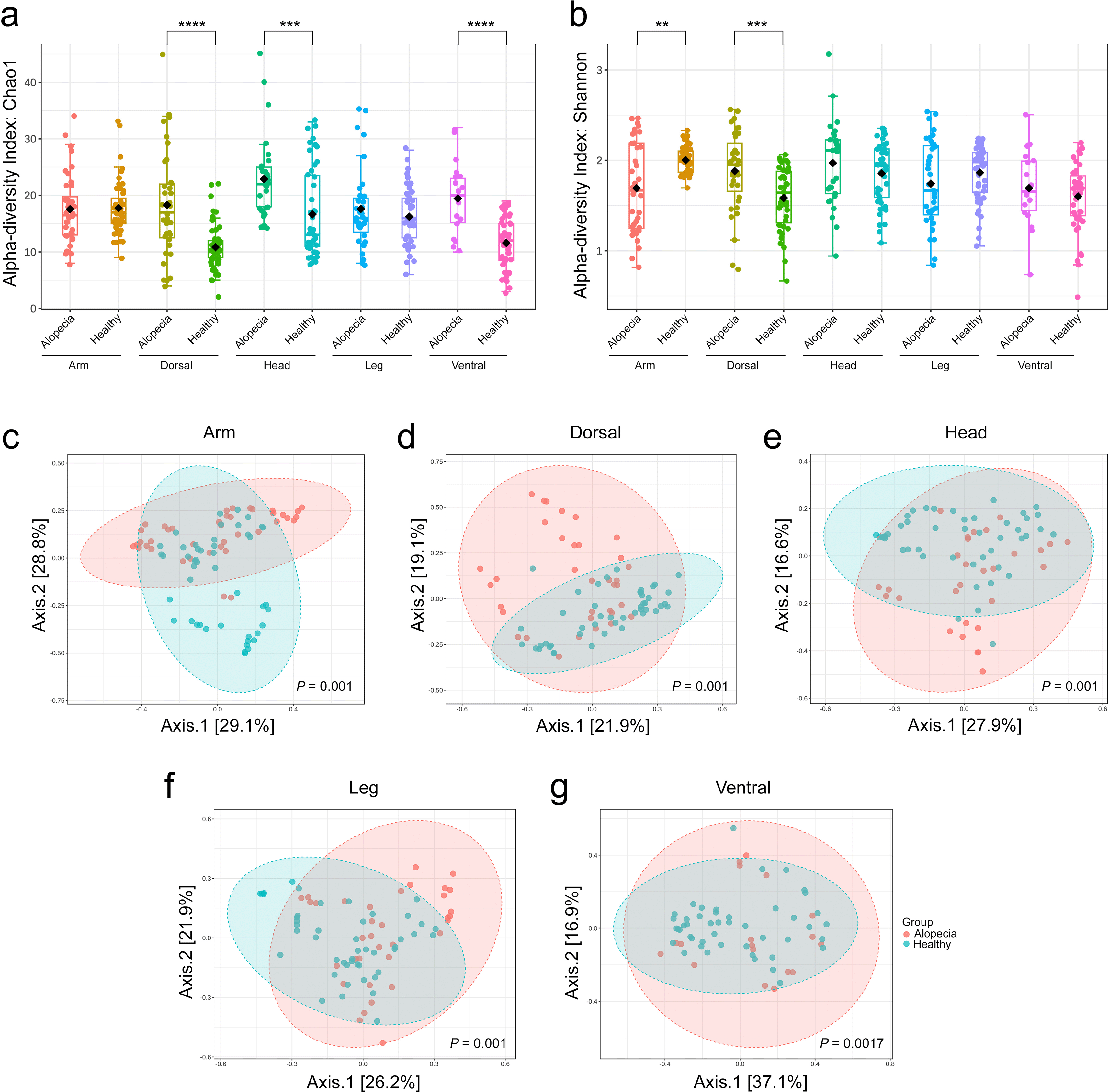
The alpha diversity of skin fungi from different body parts in healthy and alopecic cynomolgus macaques is presented in a) Chao1 index and b) Shannon index using the Kruskal–Wallis test (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). Different colors represent each group. The beta diversity is presented in a multidimensional scaling plot from Principal Coordinates Analysis (PCoA) based on Bray-Curtis dissimilarity with statistical comparisons of microbiome profiles performed using PERMANOVA (P < 0.05) of c) arm, d) dorsal, e) head, f ) leg, and g) ventral areas. The red is the alopecic group, and the blue is the healthy group.
The top two dominant phyla of fungi in the arm, dorsal, head, leg, and ventral areas were Basidiomycota and Ascomycota (Supplementary Figure S2a, c, e, g, and i). The most abundant genera were presented in Supplementary Figure S2b, d, f, h, and j. The relative abundance of fungi species in the skin is shown in Figure 2a-2e. The highest abundance of the species level indicated that Haglerozyma chiarellii was abundant in the arm (23 ± 19%), dorsal (22 ± 20%), and leg (22 ± 17.7%), and Apiotrichum montevideense was abundant in the head (11.8 ± 6.4%) and ventral (27.3 ± 18.2%) areas. Other species in the five body parts had an average abundance lower than 14%, as shown in Table 2.
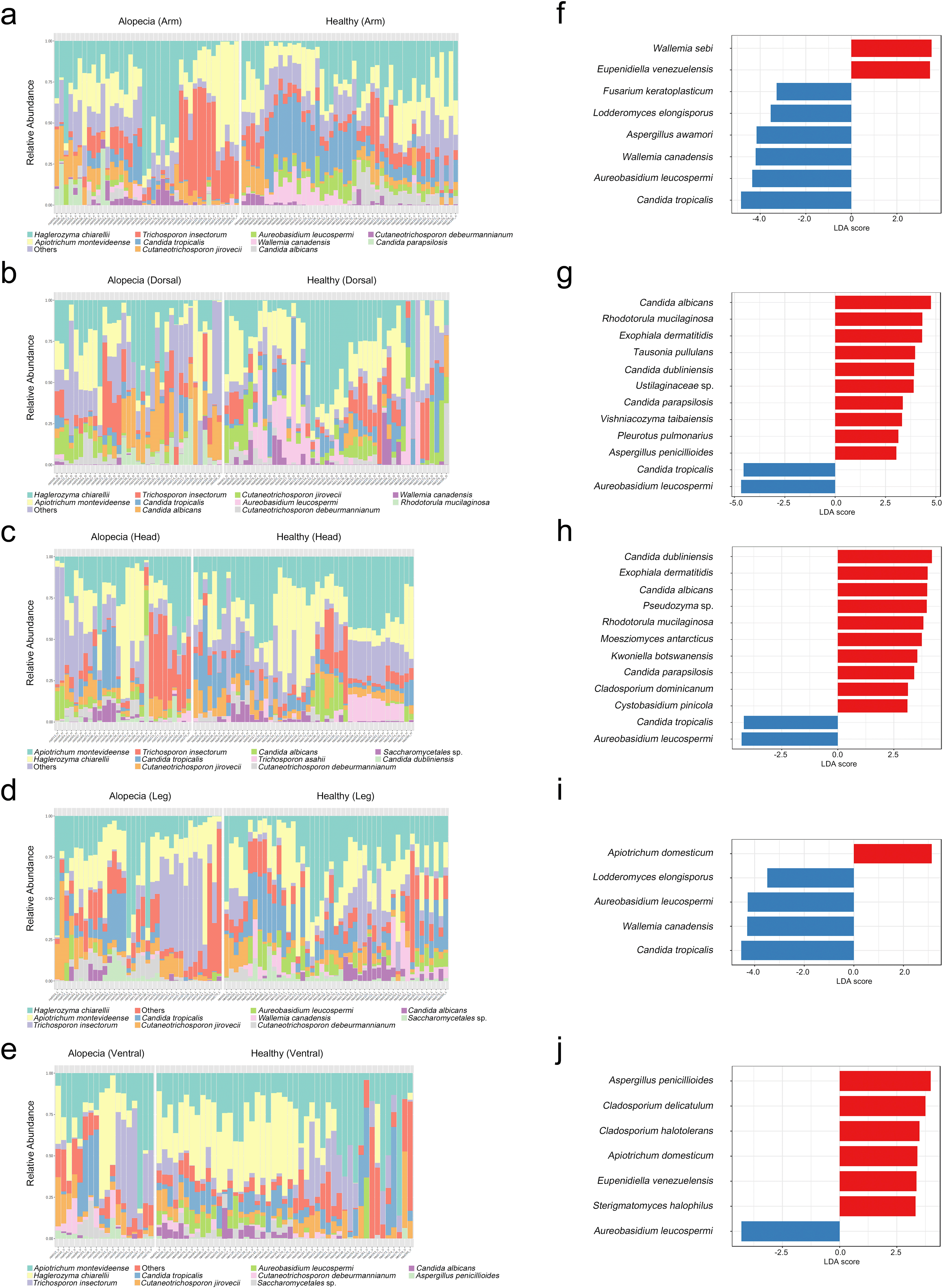
The taxonomic compositions of the skin fungi species in healthy and alopecic macaques are presented in bar graphs with the relative abundance (%). Different colored bars represented the different skin fungal taxa. LEfSe showed the most differentially abundant taxa in the five areas from alopecic (red) and healthy (blue) with the threshold of Linear discriminant analysis (LDA) score >3 and P < 0.05 (Kruskal-Wallis test). The bar graph showed a-b) arm, c-d) dorsal, e-f ) head, g-h) leg, and i-j) ventral areas.
LEfSe analysis identified significant differences in skin fungi between the two macaque groups across five body regions ( Figure 2f-2j). The bar graph showed different taxonomic levels, based on the Kruskal-Wallis test (LDA score >3, P < 0.05). For the alopecic group, LEfSe analysis demonstrated that the top abundance of fungi in the arm area was Wallemia sebi and Eupenidiella venezuelensis. The dorsal area was highly found Candida albicans, Rhodotorula mucilaginosa, Exophiala dermatitidis, Tausonia pullulans, Candida dubliniensis, Ustilaginaceae sp., Candida parapsilosis, Vishniacozyma taibaiensis, Pleurotus pulmonarius, and Aspergillus penicillioides. Furthermore, the dominant fungi in the head area were Candida dubliniensis, Exophiala dermatitidis, Candida albicans, Pseudozyma sp., Rhodotorula mucilaginosa, Moesziomyces antarcticus, Kwoniella botswanensis, Candida parapsilosis, Cladosporium dominicanum, and Cystobasidium pinicola. The leg area was uniquely found with Apiotrichum domesticum. Lastly, the prevalent species in the ventral area were Aspergillus penicillioides, Cladosporium delicatulum, Cladosporium halotolerans, Apiotrichum domesticum, Eupenidiella venezuelensis, and Sterigmatomyces halophilus.
On the other hand, LEfSe analysis showed significant skin fungi for the healthy group, with the top abundance in the arm area being Candida tropicalis, Aureobasidium leucospermi, Wallemia canadensis, Aspergillus awamori, Lodderomyces elongisporus, and Fusarium keratoplasticum. Additionally, the dorsal area and the head area were found to be Aureobasidium leucospermi and Candida tropicalis, which are the prevalent species in both areas. The leg area was highly found Candida tropicalis, Wallemia canadensis, Aureobasidium leucospermi, and Lodderomyces elongisporus. Lastly, the only dominant species in the ventral area was Aureobasidium leucospermi.
This is the first study to investigate the association between the skin mycobiota and alopecia in cynomolgus macaques. Full-length ITS sequencing using Oxford Nanopore Technologies was utilized to explore fungal community profiles. The alpha diversities of the skin fungi in five body parts between alopecic and healthy macaques were significantly different. Moreover, the Chao1 index of skin fungi of the alopecic group was increased compared to the healthy group. Previous studies reported that dysbiosis refers to an imbalance in the skin microbiome, where the typical microorganism composition is disrupted. This imbalance may involve the overgrowth or depletion of specific microbial species, leading to various dermatological conditions.29,30 The beta diversity showed that the skin mycobiota profiles of five areas between healthy and alopecic groups were significantly different. This suggests a strong relationship between the skin mycobiota and alopecic conditions. The relative abundance presented in the skin mycobiota varied among groups. Therefore, the skin mycobiome composition may be affected by possible factors such as environmental exposure, grooming behavior, or social transmission.5,31 This is supported by Sarkar et al, social transmission of microorganisms provides relationship evidence, indicating that macaques living in the same group have more similar microbiomes than macaques who live in different groups.31
For the skin fungi, Candida spp., Apiotrichum spp., Cutaneotrichosporon spp., and Trichosporon spp. in alopecic and healthy cynomolgus macaque skin were also opportunistic pathogens in humans.32 In addition, Candida spp. was a pathogenic fungal species found in the lesional skin of the pre-treatment atopic dermatitis patient.33 Trichosporon spp.was presented in the mycobiota of the epidermis, mucous membranes, and nails of mammals, can induce trichosporonosis.34
The differential abundance analysis demonstrated that significant skin fungi in the alopecic cynomolgus macaques were Wallemia sebi, which was uniquely found in the arm area. Candida albicans, Rhodotorula mucilaginosa, Exophiala dermatitidis, and Candida parapsilosis were highly observed in the dorsal and head areas. Moreover, Aspergillus penicillioides was prevalent in the dorsal and ventral areas. Pseudozyma sp., Moesziomyces antarcticus, and Cladosporium dominicanum were uniquely found in the head area. Apiotrichum domesticum was top abundant in the leg and ventral areas. Lastly, Cladosporium halotolerans was uniquely ventral. In contrast, significant skin fungi in the healthy cynomolgus macaques were Aureobasidium leucospermi, which were highly discovered in the five body parts. Furthermore, skin fungi in the healthy group were Candida tropicalis, which were found in the arm, dorsal, head, and leg areas. Lastly, Aspergillus awamori and Fusarium keratoplasticum were uniquely detected in the arm area.
For the alopecic group, Candida albicans was a dimorphic fungus that frequently colonized the oral cavity and caused opportunistic infection in the immunosuppressed.35 It was also significantly prevalent in psoriatic patients36 and in skin inflammation mice.37 As previously demonstrated, Candida parapsilosis was an important human pathogen.38 In Tinea capitis patients’ infections previously were caused by Rhodotorula mucilaginosa with immunosuppression. Rhodotorula must be regarded as a potential human pathogen.39 Exophiala dermatitidis, a black fungus that can be isolated from natural environments, was frequently reported as a human pathogen that has been isolated from subcutaneous tissue and the epidermis.40 Aspergillus penicillioides was a prevalent indoor xerophilic fungus and a potential cause of respiratory conditions in humans.41 For Wallemia sebi, this fungus was also prevalent in in-house dust. It was well-established that the health consequences of chronic exposure to mold and moisture were linked to various inflammatory compounds and human allergens,42 as reported previously. Furthermore, Pseudozyma spp. was classified as an environmental yeast. However, it has also been identified as an uncommon human pathogen in immunocompromised patients.43 Previously, Moesziomyces spp. have been identified as plant and human pathogens in the phyllosphere.44 Cladosporium dominicanum was previously isolated from fruit surfaces.45 The genus also encompassed common endophytes, plant pathogens that frequently induced leaf blotches or other lesions, as well as hyperparasites of other fungi.46 The previous study showed Cladosporium halotolerans was a saprobic and indoor fungus that was infrequently associated with human infections.47 Lastly, Apiotrichum spp. was previously one of the soil-associated species.48 For the healthy group, Candida parapsilosis was separated from skin and hair in healthy pigs and was widely distributed in the environment, including human skin, vagina, mouth, and digestive tract. It has the potential to become pathogenic following modifications to the host’s immune system, leading to localized infections and, in some cases, systemic disease.49 Aureobasidium species, while recognized as pathogens of plants and humans, are also acknowledged for their beneficial roles as plant growth promoters and biological control agents in a variety of crops and fruits, including grapes, berries, apples, pears, citrus, tomatoes, peaches, and strawberries.50 Aspergillus awamori was used to biotransform soybean extract, as the enzymatic filtrate of this fungus exhibited significant nutricosmetic potential, demonstrating both safety and the ability to effectively enhance collagen-I production in human fibroblasts.51 Although Fusarium keratoplasticum was a frequently encountered species in human infections and was implicated in a range of diseases affecting both immunocompetent and immunocompromised individuals.52 These suggest that the emergence of fungi as significant causes of human disease has been notably prevalent among immunocompromised individuals.38 Moreover, most human fungal pathogens were frequently discovered as part of natural environments or as pathogens that were already associated with the host, either as commensals or as pathogens.32
Therefore, the onset of autoimmune diseases that affect the skin may be influenced by disruptions in the microbial communities on the skin.53 Most of the previous studies in humans and NHPs utilized short-read ITS sequencing, which restricts fungal categorization predominantly to the genus level. Our study recommends full-length ITS sequencing for the advanced exploration of mycobiota associated with alopecia in macaques, potentially enhancing fungal identifications. However, ONT provided a greater resolution in identifying fungal species.
Our finding showed the difference in skin mycobiota between alopecia and healthy macaques implies that the grooming behavior of macaques, which involves physical contact to clean each other, may influence the diversity of fungi found in various body parts because the presence of fecal and oral microorganisms on the skin suggests that grooming behavior in macaques facilitates the exchange of microbes on the skin. This is supported by Bernstein et al., who reported that overgrooming or barbering is the most prevalent cause of focal alopecia.9 Moreover, the exchange of microbiomes on the skin may be influenced by physical contact during grooming, such as through saliva or fecal residues.31 Therefore, the development of alopecia may be associated with this microbial exchange. The skin microbiome can be disrupted when these factors encounter the skin in an environment conducive to colonization, resulting in the proliferation or imbalance of specific microorganisms. Therefore, it may decrease overall commensal strains.36 This dysbiosis may contribute to alopecia, as microorganisms can function as opportunistic pathogens, causing inflammation or infections that damage hair follicles, all of which may be associated with alopecia.54
In summary, an association was found between the skin mycobiota and alopecic condition in cynomolgus macaques. The presence of these fungi on the skin may imply an imbalance or disruption in the typical microbial ecosystem, which could contribute to skin conditions such as alopecia or skin inflammation. Furthermore, some fungi are common components of the skin mycobiota; however, they can become pathogenic when the skin barrier is compromised or there is skin dysbiosis. It has the potential to result in hair loss or skin infections. In the future, these findings may prove beneficial for the therapeutic management strategies of animals in primate facilities that are accredited by national standards or AAALAC International, as well as for biomedical research.
The study protocol was approved by the Animal Care and Use Committee of the National Primate Research Center of Thailand Chulalongkorn University (NPRCT-CU) (Protocol Review No. 2375015).
N.P. conducted the study design, carried out the laboratory work in 16S and ITS sequencing, and wrote the manuscript. S.V. and P.K. performed the analysis. P.C. carried out the ONT support. K.P. and T.K. collected the macaque samples for analysis. V.S. advised on the knowledge. A.K. edited the figures for the manuscript. S.M. supervised the project, edited the manuscript, and provided the macaque samples. S.P. devised the study design, supervised the study, and edited the manuscript. All authors reviewed and approved the final manuscript.
The sequencing datasets used in this study are publicly accessible through the NCBI Sequence Read Archive (SRA), BioProject ID: PRJNA1208253; https://dataview.ncbi.nlm.nih.gov/object/PRJNA1208253?reviewer=vjdlb81ful5hap5k861uar4q3l (Natthanit et al., 2025).
Figshare: Analysis of skin mycobiota associated with alopecia in captive cynomolgus macaques (Macaca fascicularis) based on Oxford Nanopore Technologies. https://doi.org/10.6084/m9.figshare.30452765 (Natthanit et al., 2025).
This project contains the following extended data:
• Supplementary Table 1. (The table shows the demographics of cynomolgus macaques housed at NPRCT-CU that enrolled in this study.)
• Supplementary Figures. (Supplementary Figures show the rarefaction curve and the fungal composition profile at the phylum and genus levels.)
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Figshare: ARRIVE Checklist for “Analysis of skin mycobiota associated with alopecia in captive cynomolgus macaques (Macaca fascicularis) based on Oxford Nanopore Technologies”. https://doi.org/10.6084/m9.figshare.30164326 (Natthanit et al., 2025).
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
The authors are grateful to thank all staff from the National Primate Research Center of Thailand, Chulalongkorn University (NPRCT-CU), for their assistance in sample collection from cynomolgus macaques. We would also like to thank the Center of Excellence in Systems Biology, Faculty of Medicine, Chulalongkorn University, for supporting the laboratory work and performing the analysis.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)