Keywords
dermatological diagnosis, artificial intelligence, convolutional neural networks, deep learning, ResNet152, Streamlit, software development, medical image classification
This article is included in the Artificial Intelligence and Machine Learning gateway.
This article is included in the AI in Medicine and Healthcare collection.
Artificial intelligence (AI)-assisted dermatological diagnosis has become increasingly important due to its ability to support clinical decision-making and reduce diagnostic variability among specialists. This paper presents DERMOSAN, an interactive software tool developed in Streamlit, based on a convolutional neural network (CNN), designed for the automatic classification of dermatological diseases using clinical images.
A public Kaggle dataset with 27,153 dermatological images distributed across ten clinical categories was used. The model was built on the ResNet152 architecture with transfer learning, implemented in TensorFlow and Keras. The images were divided into training (80%), validation (10%), and test (10%) subsets, applying class weighting to mitigate data imbalance. The tool integrates a Streamlit-based graphical interface that enables real-time image upload, local processing, and probabilistic diagnosis visualization.
The model achieved 95% accuracy in training, 83% in validation, and 92.3% in testing. The confusion matrix showed better performance in the most representative classes. The developed interface allows for automatic diagnosis along with confidence scores and complementary clinical suggestions, facilitating quick and reproducible visual interpretation.
DERMOSAN is a stage of software development aimed at integrating deep learning models into accessible clinical environments. Although the system does not include clinical validation, it represents a step toward the creation of reproducible, open-source tools for AI-assisted diagnosis.
dermatological diagnosis, artificial intelligence, convolutional neural networks, deep learning, ResNet152, Streamlit, software development, medical image classification
Over the past decade, artificial intelligence (AI) has significantly transformed the field of healthcare due to its ability to analyze large volumes of clinical data and assist medical professionals in decision-making.1 In dermatology, this transformation has been particularly relevant, given that the diagnosis of skin diseases depends largely on visual observation and the experience of the specialist, which can lead to diagnostic variability and delays in timely treatment.2 In this context, Convolutional Neural Networks (CNNs) have established themselves as one of the most promising tools for the automated classification of medical images.3
At the same time, artificial intelligence has also been successfully applied in other domains through the development of expert systems for assisted decision-making. For example, Huayta-Gómez and Pacheco4 implemented an expert system for vocational guidance, structured around a six-model approach—organizational, task and agent, knowledge, communication, design, and implementation—that optimized the process of diagnosis and self-knowledge among students. This type of methodological approach demonstrates the versatility of artificial intelligence in the design of interactive and reproducible solutions, principles that also guide the development of the present study in the context of assisted dermatological diagnosis.
Several studies have explored the application of deep learning models to support dermatological diagnosis. For example, Kadhim and Kamil5 developed a machine learning-based system for the classification of dermatological images, optimizing the clinical analysis process. Similarly, Lesaunier et al.6 and Zhang et al.7 showed that the incorporation of information technology systems in areas such as interventional radiology improves efficiency and reduces errors through the application of Lean principles.8 These advances demonstrate the potential of AI to transform medical diagnosis, although they also highlight limitations related to the accessibility and reproducibility of the tools developed.
CNNs, in particular, reproduce the human visual learning process through convolutional layers that allow complex patterns in images to be identified.9 Their application has spread to specialties such as radiology, digital pathology, and dermatology, achieving results comparable to and even superior to those of dermatologists in specific classification tasks.10 In Asia,11 demonstrated the effectiveness of deep architectures—including CNN, RNN, GAN, and LSTM—in improving real-time diagnostic accuracy. As shown in Figure 1, these types of networks have transformed the diagnostic process by complementing clinical evaluation with deep learning–based decision support systems. However, most of these developments are concentrated in environments with high technological availability and large databases, which limits their adoption in developing countries.
Source: Own elaboration.
Note: Comparison between traditional dermatological diagnosis and diagnosis assisted by convolutional neural networks (CNN).
In Latin America, the adoption of AI-based tools has advanced gradually. Tentori et al.12 reported the development of telemedicine and computer-assisted diagnosis initiatives in Brazil and Mexico, showing promising results despite persistent challenges related to technological infrastructure and access to standardized clinical data. In Peru, studies by Ponce et al.13 and Sarria et al.14 highlighted advances in medical automation, yet revealed the lack of local solutions focused on dermatological image analysis using deep learning models. This gap emphasizes the need for accessible and reproducible tools designed for academic research and clinical integration of artificial intelligence.
In response to this need, DERMOSAN, an interactive software tool in Streamlit for CNN-assisted dermatological diagnosis, was developed. Its purpose is to offer an intuitive development phase platform that allows dermatological images to be uploaded and probabilistic diagnoses to be obtained in real time. Unlike previous solutions, DERMOSAN integrates an advanced CNN architecture (ResNet152) with an interactive open-source interface, facilitating its use for academic, research, and demonstration purposes, promoting reproducibility and support for medical specialists.
In this section, we describe the study design, the origin of the data, the technical implementation of the model, and the operation of the assisted dermatological diagnosis tool.
This work corresponds to an applied study with a quantitative and methodological-observational approach, aimed at developing and validating an automated diagnostic tool.
The dataset was stratified by class into three subsets: training (80%), validation (10%), and testing (10%). To ensure reproducibility, a fixed random seed (SEED = 42) was used in all operations involving randomness (such as random image sampling and internal shuffling). This allows the partitioning and random behaviors to be consistent when running the same code with the same data and environment.
During training, the validation set was used to adjust hyperparameters and control overfitting; the independent test set was used exclusively for the final evaluation of the model’s performance and generalization.
The public dataset was obtained from Kaggle and comprises 27,153 dermatological images distributed across ten clinical categories. Table 1 summarizes the number of images per class and subset (training, validation, and test), revealing an imbalance among some categories. As illustrated in Figure 2, this imbalance becomes more evident in the bar chart, where the Melanocytic Nevi (NV) class accounts for the largest proportion of images, highlighting the non-uniform distribution among classes.
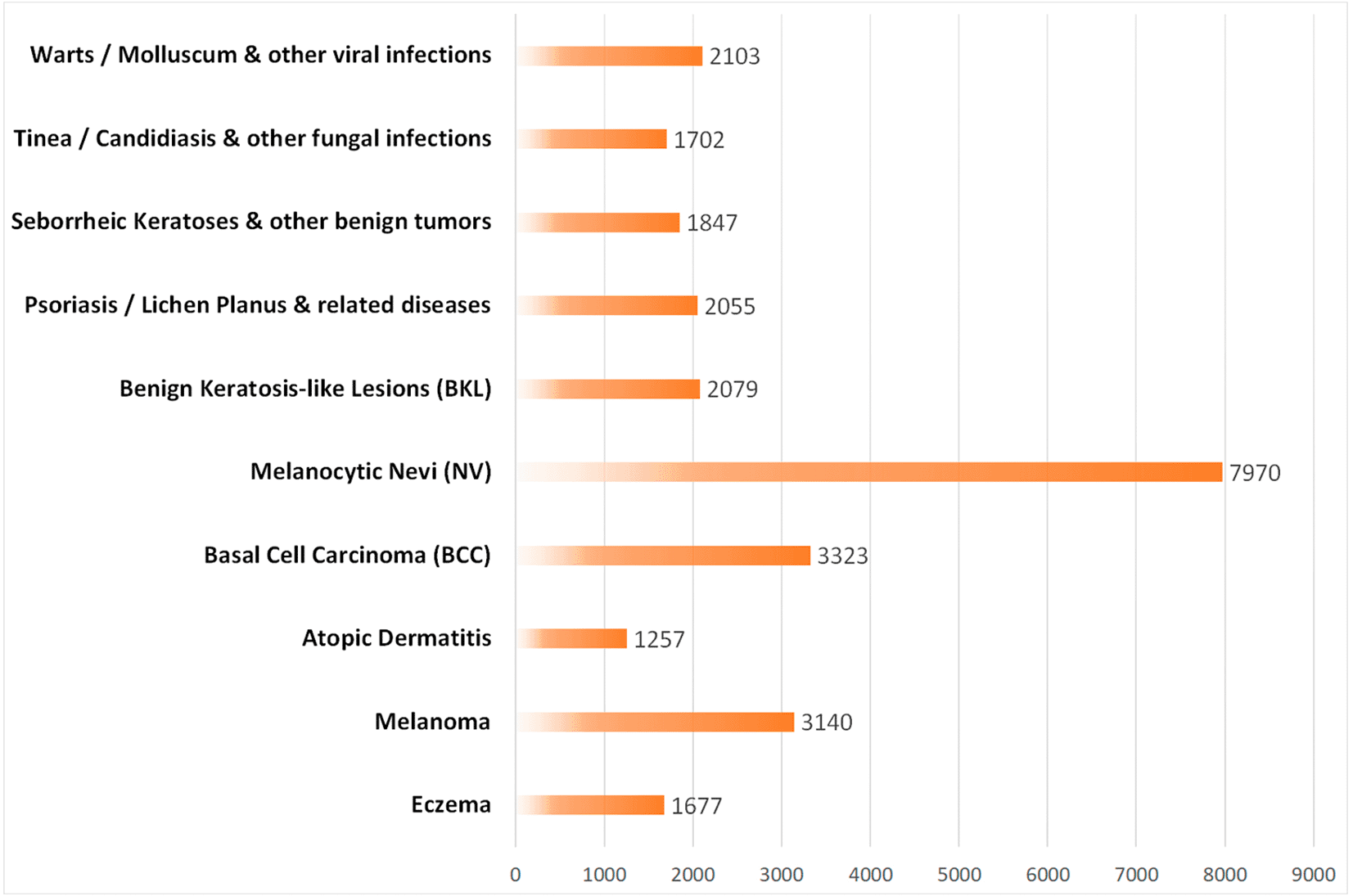
Source: Own elaboration.
Note: The Melanocytic Nevi (NV) class accounts for the largest proportion of images, indicating an imbalance.
Due to the imbalance between classes, the class_weight parameter was used during training to adjust the relative importance of each class in the error calculation. As a result, classes with fewer examples received a greater penalty when making errors, which encourages the model to pay more attention to learning those classes and not skew toward the dominant classes.
Technologies and library integration
The diagnostic tool was developed in Python 3.10, using the TensorFlow 2.15.0 and Keras 3.0 frameworks to implement the ResNet152 model using the transfer learning technique.
In addition, several complementary libraries were incorporated to strengthen the overall functionality of the system. Scikit-learn was used to calculate performance metrics and generate the confusion matrix, while Plotly, Matplotlib, and Seaborn were used for interactive visualization of loss curves, accuracy, and clinical graphs.
NumPy and Pandas enabled the manipulation and structured analysis of data, ensuring efficient statistical processing, while OpenCV and Pillow were used for the preprocessing and validation of dermatological images.
Finally, Streamlit 1.32.0 was implemented as the development environment for the graphical interface, facilitating direct interaction between the user and the predictive model, as well as immediate and reproducible visualization of diagnostic results.
Methodology in development phases
To structure the DERMOSAN development process, a phased methodological approach was adopted, inspired by the model proposed by Ramos-Miller and Pacheco,15 who implemented a five-phase methodology—comprising analysis, planning, implementation, review, and deployment—in the development of an educational web-based inventory control system. This sequential approach enhances the traceability of requirements and enables a systematic evaluation of the software throughout its development cycle.
▪ Phase 1: Data preprocessing and preparation
Images (JPEG, PNG, WEBP) are read, decoded, resized to 224×224 px, and normalized using preprocess_input from the ResNet model. Class mapping and organization into directories by class are generated. A global seed (SEED = 42) was set to ensure reproducibility in sampling and shuffling. In addition, the resulting dataset is shuffled internally and prefetched for data reading optimization.
▪ Phase 2: Stratified division and calculation of class weights
The images are partitioned into training (80%), validation (10%), and testing (10%) in a stratified manner by class. Class_weight is then calculated using the ‘balanced’ method, so that less frequent classes receive a higher weight during training, mitigating biases.
▪ Phase 3: Model design and configuration
ResNet152 is adopted with pre-trained weights from ImageNet. All layers except the last 50 are frozen for fine-tuning. Dense layers with ReLU activations, dropout = 0.35, and a final softmax layer are added. The model is compiled using the Adam optimizer and sparse categorical cross-entropy loss. ReduceLROnPlateau (factor = 0.2, patience = 2, min_lr = 1e-6) and ModelCheckpoint (saves weights at the end of each epoch) callbacks are configured.
▪ Phase 4: Training and validation
Training is run for a total of 9 epochs (or resumes from checkpoint if one exists). In each epoch, val_loss is evaluated to adjust the learning rate and save the best performing model. The optimal model is exported as best_resnet152.h5.
▪ Phase 5: Integration with the interface and local deployment
The final trained model is incorporated into an application developed with Streamlit. The application allows the user to upload images, validate them (e.g., format, resolution), process them through inference, and view the diagnosis, confidence level, and estimated risk. The system is internally modularized into components (image validation, inference, visualization) to facilitate maintenance and extensibility.
Graphical interface and processing flow
The application, developed with Streamlit, features an interactive interface that allows dermatological images in JPG, PNG, or WEBP format to be uploaded for automated analysis. Once the image is uploaded, the system runs an internal validation process that evaluates aspects such as sharpness, lighting, and resolution, ensuring that the images are suitable for assisted diagnosis.
The validated image is then processed by the pre-trained ResNet152 model, which generates an automatic classification with the main diagnosis, the confidence level, and an estimate of the associated clinical risk. The results are presented in a visual panel that combines quantitative and graphical information, facilitating the interpretation of predictions by the user.
Figure 3 schematically illustrates the tool’s processing flow, from image loading to final diagnosis generation.

Source: Own elaboration.
Note: The diagram illustrates the initial stages of the system, from image loading by the user to the generation of the automated diagnosis.
This application is configured as an interactive and reproducible clinical support tool that contributes to the automated analysis of dermatological images without replacing the diagnostic work of the specialist.
Minimum system requirements
The application was designed to run locally on personal computers, without the need for an internet connection or cloud infrastructure. The minimum requirements for its operation are as follows:
▪ Operating system: Windows 10 or Ubuntu 20.04 or higher.
▪ Processor (CPU): Intel Core i5 or equivalent.
▪ RAM: Minimum 8 GB.
▪ Graphics processing unit (GPU): Optional, although an NVIDIA T4 (16 GB VRAM) is recommended to accelerate model inference.
▪ Software dependencies: Python ≥ 3.10, TensorFlow, Keras, Streamlit, NumPy, Pandas, and scikit-learn.
To run the application, simply open a terminal in the project directory and execute the command: streamlit run app.py
Once the environment is initialized, the interface can be viewed from a modern web browser with JavaScript enabled, by accessing the local address indicated by Streamlit (by default, http://localhost:8501).
The main metric considered was overall accuracy, calculated on the test set by comparing actual labels with model predictions. In addition, the confusion matrix was analyzed, which allowed us to identify error patterns between categories, as well as the loss and accuracy curves recorded during the training and validation phases.
The study used only a public, previously anonymized dataset of dermatological images (Kaggle), in compliance with the repository’s terms of use.
As it did not contain any identifiable patient information, approval by an institutional ethics committee was not required. The system was developed for academic purposes and is a diagnostic support tool, not a substitute for the medical judgment of a specialist.
The model developed achieved an accuracy of 95% in training, 83% in validation, and 92.3% in testing, reflecting the technical performance achieved in the development stage of the DERMOSAN software. These values indicate solid model performance, with slight differences between training and validation attributable to class imbalance.
Figure 4 shows the confusion matrix obtained in the test set. Superior performance is observed in classes with a greater number of images, such as Melanocytic Nevi (NV) and Basal Cell Carcinoma (BCC), while minority categories (Eczema, Atopic Dermatitis) show more classification errors.

Source: Own elaboration.
Note: The model performs better in classes with a larger number of images, while minority categories show more classification errors.
Figure 5 shows the evolution of loss during training and validation. A progressive reduction is observed in the training set, while in validation the values remain relatively high, indicating the presence of overfitting.
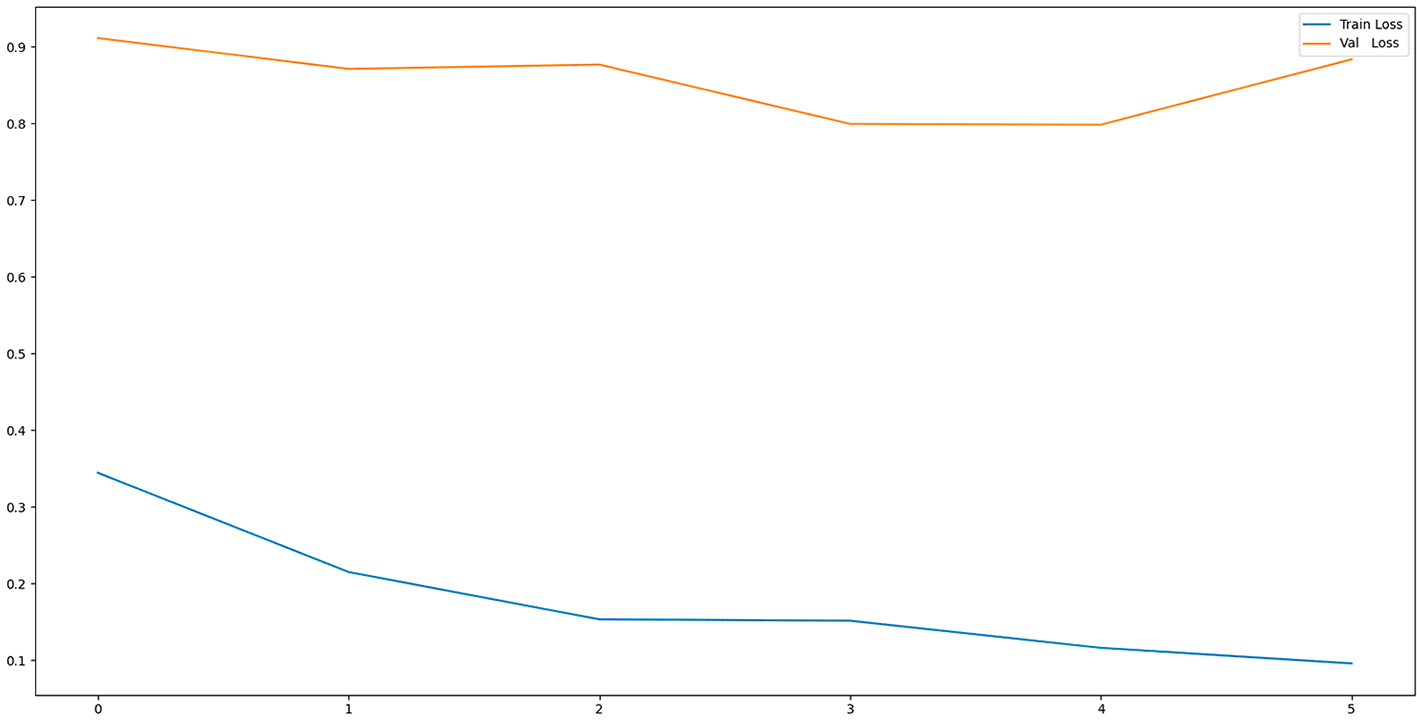
Source: Own elaboration.
Note: Loss decreases steadily during training, while remaining high during validation, indicating overfitting.
Figure 6 shows the accuracy curve. The model achieved over 95% accuracy in training, while in validation it stabilized at around 83%. This difference is related to the difficulty of adequately generalizing in classes with lower representativeness.

Source: Own elaboration.
Note: The model achieved >95% in training and ~83% in validation. The gap reflects generalization limitations derived from class imbalance.
Figure 7 shows the graphical interface developed in Streamlit, which allows individual images to be uploaded and the model’s suggested diagnosis to be obtained in real time. The application displays the main diagnosis with its confidence level, as well as the most likely differential diagnoses.
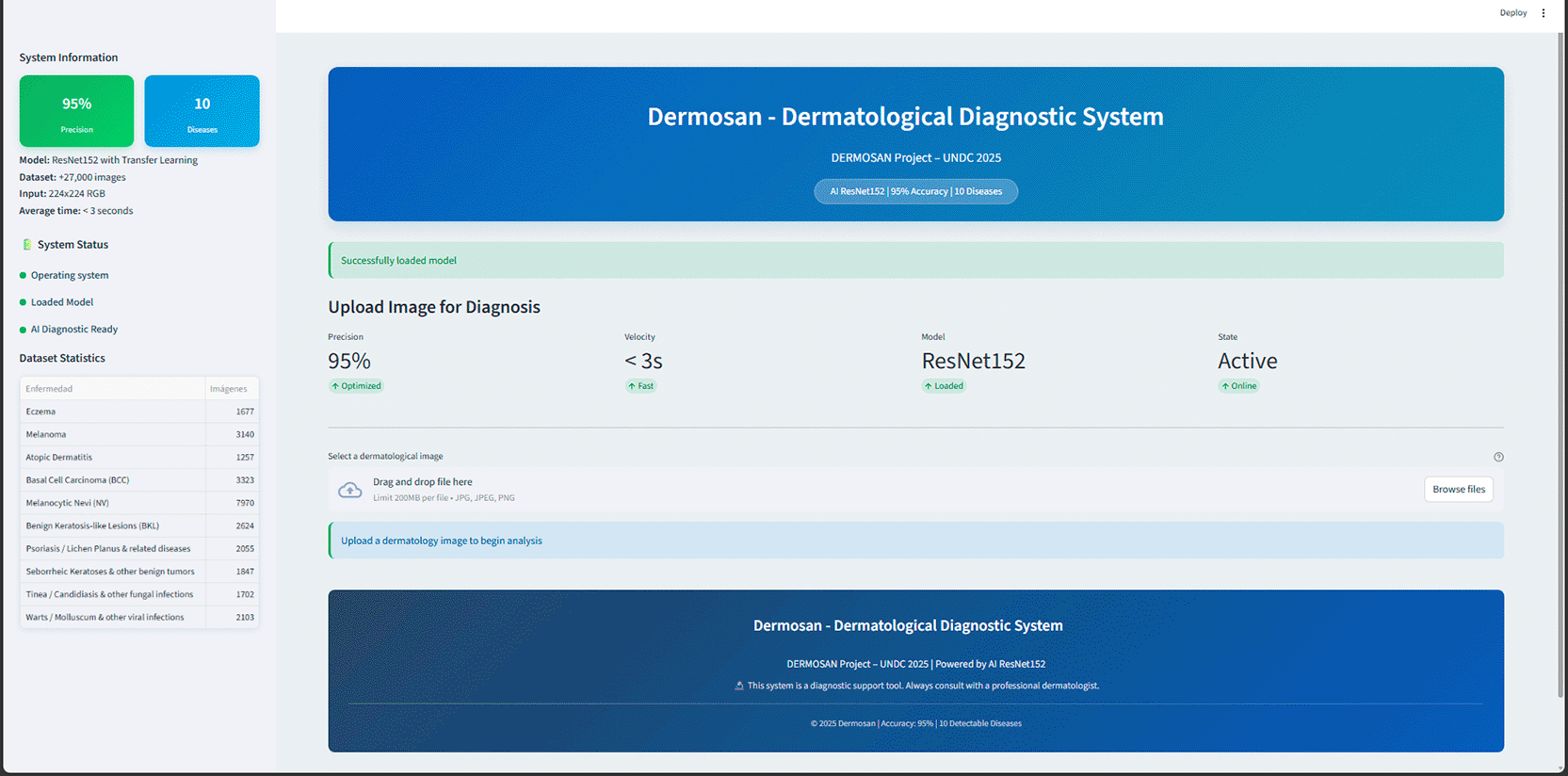
Source: Own elaboration.
Note: The tool allows you to upload an image, display the most likely diagnosis with its confidence level, and present differential diagnoses.
Finally, the overall performance of the model on the test set is shown in Figure 8, where a total accuracy of 92.3% was achieved when evaluating the 2,723 test images. This result confirms the model’s ability to generalize on unseen data, maintaining a consistent relationship with the training and validation metrics.

Source: Own elaboration.
Note: The model achieved an accuracy of 92.3% on the test set (2723 images), demonstrating good generalization ability.
Although additional metrics such as precision, recall, or F1-score were not calculated, their inclusion is recommended in future work to evaluate the balance of the model, especially in minority classes. During training, it was observed that the differences in performance between training and validation remained consistent from the early epochs, suggesting that the model learned quickly and that class imbalance limits its generalization ability in less represented classes.
To evaluate the practical functioning of the developed system, controlled tests were performed in the local version of the Streamlit environment. These tests simulated real clinical use scenarios, with the aim of verifying the model’s ability to process dermatological images, generate diagnoses, and issue interpretive recommendations.
In Case 1, the user accesses the graphical interface and selects a dermatological image in JPG, PNG, or WEBP format. The platform displays a preview and performs an automatic quality check, evaluating parameters such as sharpness, lighting, and minimum resolution requirements. If the image does not meet the established criteria, the system notifies the user to replace it before analysis. Once validated, the interface confirms that the image is suitable for diagnosis ( Figure 9).
In Case 2, the validated image is sent to the pre-trained ResNet152 model with transfer learning, which processes the sample locally using the weights stored in best_resnet152.h5. Based on this inference, the system classifies the skin lesion into one of ten available clinical categories and displays the diagnosis with its confidence level.
For example, one image was classified as Benign Keratosis-like Lesions (BKL) with a confidence of 98.1%, while other categories such as Basal Cell Carcinoma (BCC) had lower probabilities. The result is displayed in a bar chart accompanied by interpretive text that facilitates understanding of the analysis ( Figure 10).
In Case 3, the system expands the result with an advanced clinical analysis, which includes risk assessment, diagnostic comparison with the three most likely classes, and a confidence distribution graph ( Figure 11). Subsequently, the user is presented with a detailed clinical panel that includes a description of the lesion, the risk level, medical recommendations, and a cautionary note clarifying that the diagnosis must be confirmed by a dermatologist. This stage reinforces the tool’s function as a clinical diagnosis support system, not as a substitute for professional judgment ( Figure 12).

Source: Own elaboration.

Source: Own elaboration.
Together, the cases described represent the entire workflow of the system, from image loading and validation to clinical interpretation of the final result. This workflow demonstrates the tool’s ability to assist in dermatological analysis in a reproducible, understandable, and medically oriented manner, consolidating its potential application in clinical environments for AI-assisted diagnosis.
The implementation of the CNN-based model (ResNet152 with transfer learning) demonstrated a notable improvement in the automatic classification of dermatological conditions, achieving accuracies of 95% for training, 83% for validation, and 92.3% for testing. These results indicate that deep learning models can effectively complement dermatological diagnosis by reducing the variability associated with human clinical judgment. The obtained performance aligns with that reported by Liu et al.,16 who developed a multi-classifier architecture for skin lesion recognition using multi-scale attention mechanisms, achieving an accuracy of 88.2% on the HAM10000 dataset. Similarly, Musthafa et al.17 achieved 97.78% using an optimized CNN, while Shetty et al.18 reported 95.18% with a cross-validation strategy, reinforcing the effectiveness of deep convolutional architectures combined with data augmentation techniques.
The proposed model performed better in classes with higher representativeness, such as Melanocytic Nevi and Basal Cell Carcinoma, which is consistent with the findings of Winkler et al.,19 who demonstrated that CNNs can achieve diagnostic sensitivities comparable to those of dermatologists. This trend has been repeatedly observed in dermatology, where CNN-based models have increased the diagnostic accuracy of specialists, supporting their value as clinical decision-support tools. However, minority classes such as Eczema and Atopic Dermatitis showed higher error rates, confirming the negative impact of data imbalance on model generalization—an issue also highlighted in the systematic reviews of Brinker et al.20
The use of the class_weight parameter partially mitigated the imbalance effects, improving training stability and reducing overfitting. This approach is consistent with that of Shetty et al.,18 who emphasized the importance of rebalancing strategies to enhance model robustness. In addition, integrating a Streamlit-based graphical interface enabled the model to be deployed in an interactive, accessible, and reproducible environment, facilitating real-time image analysis and the visualization of results with interpretable confidence levels. This development aligns with the recommendations of Esteva et al.,21 who underscored the need to adapt CNN-based diagnostic systems to everyday clinical workflows.
The visual outcomes obtained through the confusion matrix and training curves indicated stable and predictable performance, with deviations primarily attributable to the dataset composition. These findings are consistent with those of Liu et al.,16 who demonstrated that biases in data distribution directly affect validation metrics. Overall, the proposed tool demonstrates the feasibility of combining deep learning and accessible visual environments to support dermatological diagnosis.
The main limitations of this study include class imbalance, dependence on a single public dataset, the absence of complementary evaluation metrics such as precision, recall, and F1-score, and the lack of direct clinical validation with real cases.
For future work, we propose expanding the dataset with local samples, performing multicenter external validations, and exploring explainable AI (XAI) approaches to improve model transparency and clinician trust in real-world applications.
This study confirmed the effectiveness of convolutional neural networks (CNNs)—specifically the ResNet152 architecture with transfer learning—as a support tool for the automated diagnosis of dermatological conditions. The model achieved accuracies of 95% in training, 83% in validation, and 92.3% in testing, demonstrating strong classification performance and reproducibility under controlled conditions.
The integration of a Streamlit-based interface enabled the model to be deployed in a practical and interactive environment, facilitating its use by professionals and its potential adaptation to clinical workflows. This development demonstrates the feasibility of linking deep learning models with accessible visual applications, enhancing the interpretability of results and supporting medical decision-making.
Nevertheless, the study has limitations related to class imbalance and reliance on a single public dataset, which may affect the generalization capability of the model in broader clinical contexts. Future research should address these limitations by incorporating more representative datasets, conducting multicenter external validations, and incorporating additional evaluation metrics such as precision, recall, and F1-score for a more comprehensive assessment of model performance.
Overall, this research represents an initial step toward the practical integration of artificial intelligence in dermatology, underscoring the potential of CNN-based tools to complement clinical diagnosis. Under professional supervision, such systems could significantly contribute to improving diagnostic accuracy and efficiency in dermatological care.
Software available from: ---
Source code available from: https://github.com/tavoofg/dermosan-dermatology-cnn
Archived source code at time of publication: https://doi.org/10.5281/zenodo.1728110223
License:* https://opensource.org/license/MIT
The dataset used in this study corresponds to the “Skin Diseases Image Dataset” originally published by I. Hossain on Kaggle.22
Original source: https://www.kaggle.com/datasets/ismailpromus/skin-diseases-image-dataset 22
The dataset is openly accessible; however, all data files are © Original Authors, as stated by the original source. No modifications were made to the dataset for this research, and therefore it is not redistributed in this publication.
Instead, users should access the dataset directly from its original source to ensure proper licensing and attribution.
License for underlying data
As indicated by the creators, the dataset is provided under the license specified on the original Kaggle page (“Data files © Original Authors”). This license does not permit redistribution; therefore, only the original source is linked.
Special recognition is given to the Universidad Nacional de Cañete (UNDC), as well as to the teachers, students, and collaborators who, with their commitment and guidance, made it possible to develop this scientific article within the framework of the academic training received.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)