Keywords
Transcriptomics, Bioinformatics, Vaccination, Inoculation, Respiratory viral infection, Influenza viruses, COVID, Respiratory syncytial viruses (RSV), Human rhinoviruses (HRV) Whole Blood, PBMC.
Our understanding of the human immune system’s response to viral respiratory tract infections (VRTIs) and vaccines, including the molecular mechanisms and correlates of protection, remains incomplete. Extensive transcriptomic data from inoculation and vaccination studies have been deposited in publicly available databases. However, these studies are often separate and difficult to locate.
To bridge this research gap, we have systematically searched and reviewed publicly available datasets from NCBI Gene Expression Omnibus (GEO), Archive of Functional Genomics Data (ArrayExpress), Immunology Database and Analysis Portal (ImmPort), and Google using targeted queries. Inoculation study queries included terms related to humans, blood, PBMCs, nasal, respiratory challenges, and respiratory viral inoculations; while vaccination study queries focused on humans, blood, PBMCs, transcriptomes, and respiratory viral vaccines or vaccinations.
This collection includes 18 datasets from inoculation of 5 respiratory viruses: H1N1, H3N2, RSV, HRV, and SARS-CoV-2 (14 from blood and 4 from nasal swabs) with 429 participants (ages 18 to 55 years) and 37 datasets from vaccination of influenza and COVID-19 with 2,084 participants (ages 0.5 to over 89 years). The duration and number of post-immunization time points range from 14 days before to 28 days after inoculation (1 to 20 time points) and from 28 days before to 360 days after vaccination (1 to 13 time points).
We provide a curated compendium of public gene expression data repositories for researchers to reanalyze transcriptomes from human whole blood, peripheral blood mononuclear cells (PBMCs), and nasal swab samples. This will facilitate studies of transcriptional responses to respiratory viral inoculation or vaccination.
Transcriptomics, Bioinformatics, Vaccination, Inoculation, Respiratory viral infection, Influenza viruses, COVID, Respiratory syncytial viruses (RSV), Human rhinoviruses (HRV) Whole Blood, PBMC.
Viral respiratory tract infections (VRTIs) are widespread globally, causing significant illnesses and hospitalizations each year.1 The importance of studying VRTIs has grown in light of the global COVID-19 pandemic.2 VRTIs involve a range of viruses, including respiratory syncytial viruses (RSV), human rhinoviruses (HRV), and influenza and coronaviruses.3 Most VRTIs lack both effective antiviral therapies and approved vaccines.4
In-depth investigation of human immune responses to inoculation and vaccination is critically needed. To facilitate further research, we have compiled a curated dataset collection, which includes transcriptomic data from the blood and nasal swabs of volunteers who underwent inoculation and vaccination, specifically for influenza, COVID-19, RSV, and HRV.
Our curated dataset collection is divided into two categories: inoculation studies and vaccination studies. The inoculation collection includes 18 datasets with 429 participants, while the vaccination collection comprises 37 datasets with 2,084 participants. All datasets were sourced from GEO, ImmPort Shared Data, and the ArrayExpress Collection at EMBL-EBI.5–7 These datasets have been organized into a publicly accessible format for download and further analysis.
The datasets were gathered using specific search queries across GEO, ArrayExpress, ImmPort, and Google search. The queries for inoculation studies included terms related to humans, blood, PBMCs, nasal, respiratory challenges, and viral inoculations. The vaccination study queries followed similar criteria, focusing on humans, blood, PBMCs, transcriptomes, and vaccines or vaccinations. The search queries used for GEO, ArrayExpress, and Google search to find inoculation studies are as follows:
(“humans”[MeSH Terms] OR “Homo sapiens”[Organism] OR Homo sapiens [All Fields]) AND ((“blood”[Subheading] OR “blood”[MeSH Terms] OR blood [All Fields]) OR PBMC [All Fields] OR PBMCs [All Fields]) AND respiratory [All Fields] AND (challenge [All Fields] OR “experimentally infected”[All Fields] OR inoculated [All Fields])
Homo sapiens AND (blood OR PBMC OR PBMCs) AND transcriptome AND respiratory AND (inoculation OR challenge OR “experimentally infected” OR inoculated)
The search queries used for GEO, ArrayExpress, and Google to find vaccination studies are as follows:
(“humans”[MeSH Terms] OR “Homo sapiens”[Organism] OR Homo sapiens [All Fields]) AND ((“blood”[Subheading] OR “blood”[MeSH Terms] OR blood [All Fields]) OR PBMC [All Fields] OR PBMCs [All Fields]) AND respiratory [All Fields] AND ((“vaccination”[MeSH Terms] OR inoculation [All Fields]) OR (“vaccines”[MeSH Terms] OR vaccine [All Fields]) OR (“vaccines”[MeSH Terms] OR vaccines [All Fields]) OR (“vaccination”[MeSH Terms] OR vaccination [All Fields]))
The majority of datasets obtained were derived from human blood, PBMCs, and nasal swabs, generated using Illumina or Affymetrix platforms or through RNA sequencing. Every dataset identified through this search was carefully curated manually. This involved thoroughly reviewing the dataset descriptions, examining the study designs, and reading through the related original articles on PubMed. Ultimately, we only included studies that involved human whole blood, PBMCs, and nasal swabs linked to VRTI inoculation or vaccination for our dataset collection. According to these criteria, we retained 18 inoculation datasets and 37 vaccine-related datasets.
The data selection process for inoculation is summarized in Figure 1a. Using GEO and ArrayExpress inoculation query, 108 studies were found with Homo sapiens as the primary organism. Google search returned 92 inoculation studies. The data selection process for vaccination is summarized in Figure 1b. GEO and ArrayExpress Vaccine query identified 82 studies where Homo sapiens is the top organism. Google search produced 89 vaccination studies. ImmPort search yielded 99 vaccination studies.

Figure 2 illustrates the distribution of participants over time in 18 inoculation studies, involving 429 participants aged 18 to 55 years, across five respiratory viruses: H1N1, H3N2, RSV, HRV, and SARS-CoV-2. There is a notable peak on day 0. The duration and number of post-immunization time points range from 14 days before to 28 days after inoculation, spanning 1 to 20 time points. Notably, 56 participants from four studies had both blood and nasal samples, and they were counted separately. Subsequent time points show smaller but consistent sample collections, with varying contributions from different pathogens. H1N1, H3N2, and HRV are the most frequently sampled pathogens, while SARS-CoV-2 and RSV contribute fewer samples to the dataset.
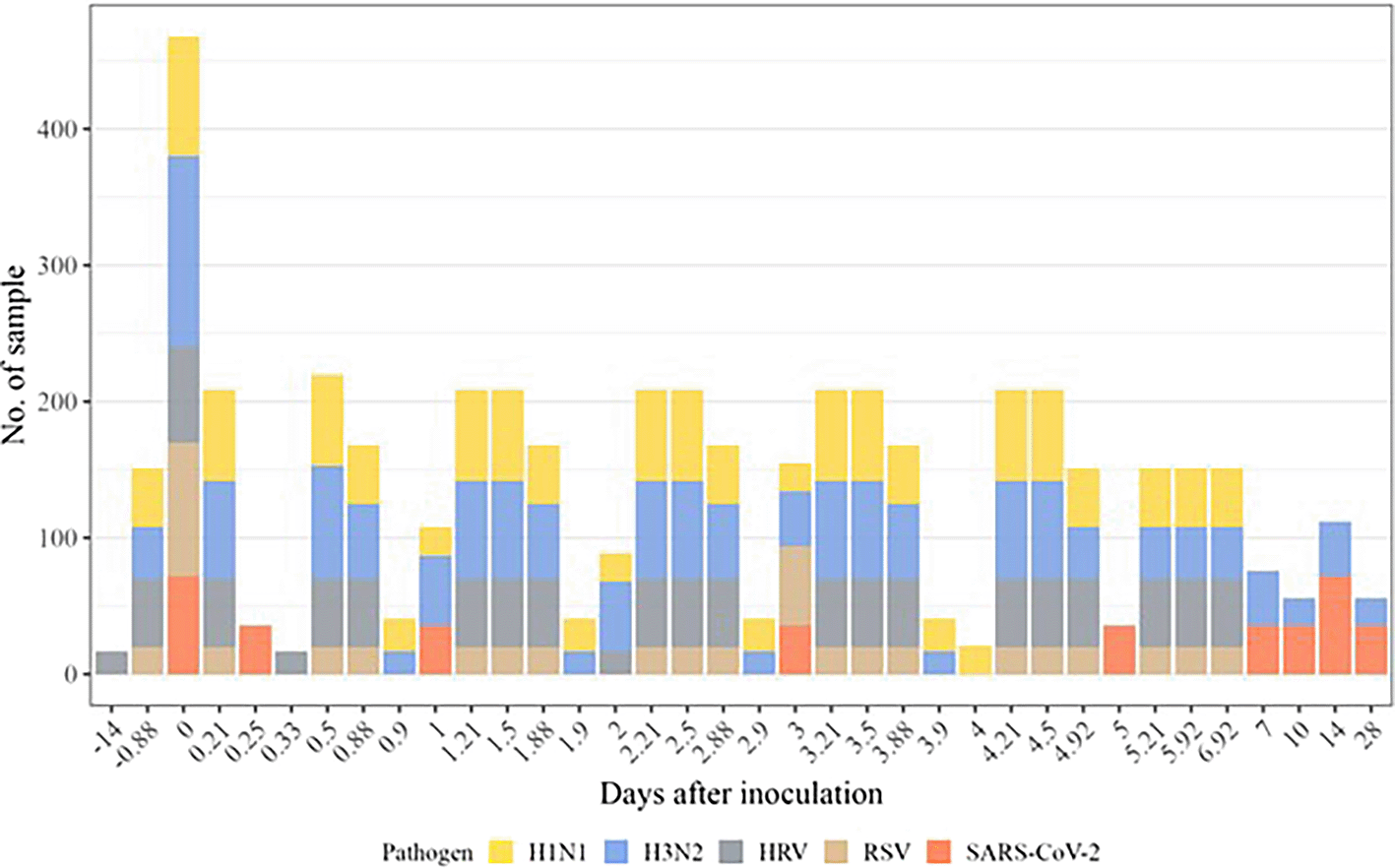
Histogram of the time points pre- and post-inoculation available in our compendium. Each virus is indicated by a different color. The height of the bars represents the number of participants with available gene expression data.
Figure 3 summarizes the distribution of 2,084 participants, aged 0.5 to over 89 years, over time in 37 vaccination studies covering COVID-19 and influenza vaccines and their types. The peak occurred on day 0, with participant recruitment enriched during the first week. The duration and number of post-immunization time points range from 28 days before to 360 days after vaccination, spanning 1 to 13 time points.
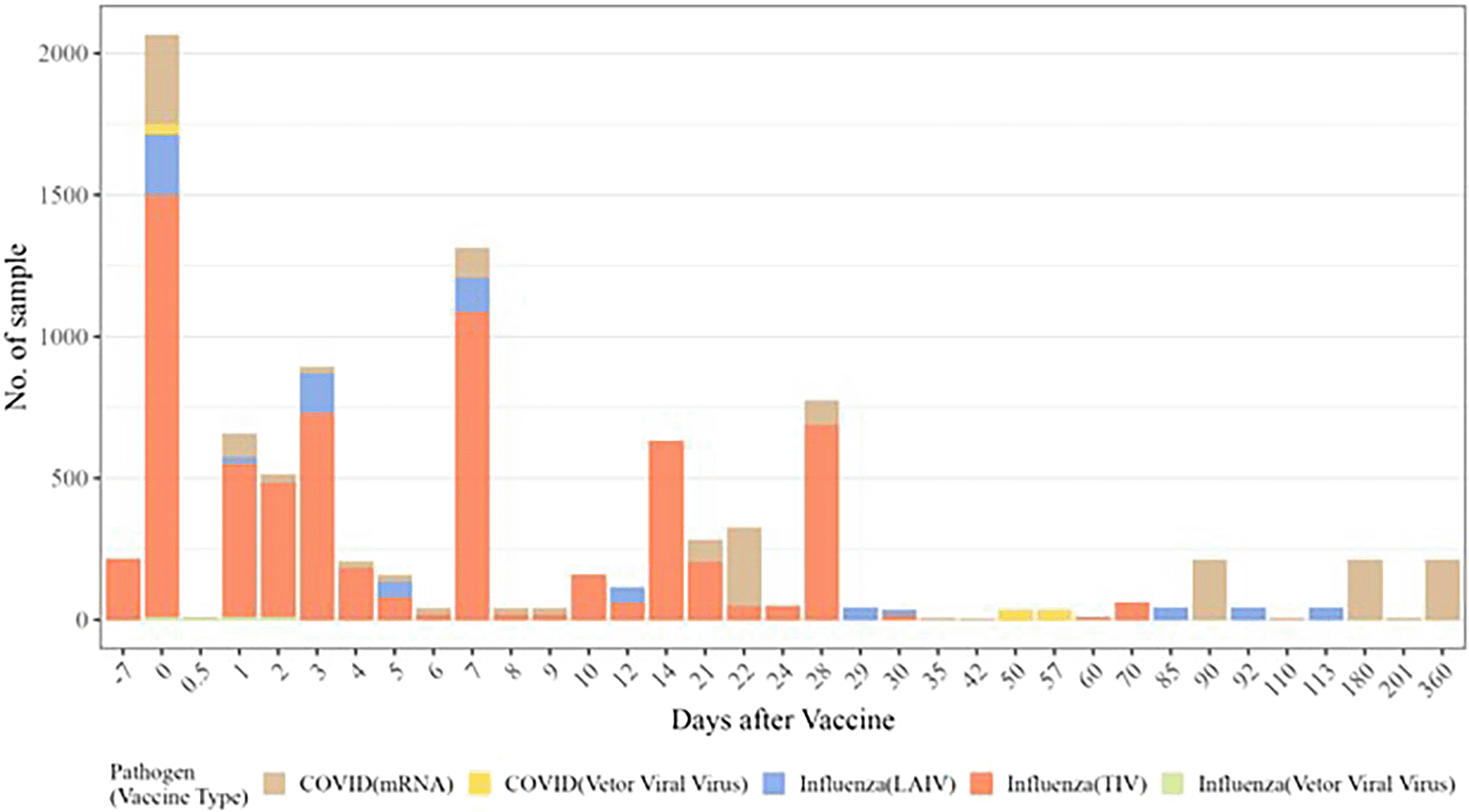
Histogram of the time points pre- and post-vaccination is available in our compendium. Each vaccine and type are indicated by a different color. The height of the bars represents the number of participants with available gene expression data.
The complete list of inoculation datasets included in our collection can be found in Table 1.
| Pathogen | Sample type | Sample size | Profiling platform | Time point | Accession number | Citation |
|---|---|---|---|---|---|---|
| HRV | Peripheral blood | 50 | Microarray (Affymetrix) | -0.88, 0, 0.21, 0.5, 0.88, 1.21, 1.5, 1.88, 2.21, 2.5, 2.88, 3.21, 3.5, 3.88, 4.21, 4.5, 4.92, 5.21, 5.92, 6.92 | GSE73072 | 8 |
| RSV | 20 | |||||
| H3N2 | 38 | |||||
| H1N1 | 43 | |||||
| HRV | Peripheral blood | 20 | Microarray (Affymetrix) | 0 | GSE17156 | 9 |
| RSV | 20 | |||||
| H3N2 | 17 | |||||
| H3N2 | Peripheral blood | 17 | Microarray (Affymetrix) | 0, 0.21, 0.50, 0.88, 1.21, 1.50, 1.88, 2.21, 2.50, 2.88, 3.21, 3.50, 3.88, 4.21, 4.50 | GSE30550 | 10 |
| H3N2 | Whole blood | 11 | Microarray (Illumina) | 0,0.50,1,2 | GSE61754 | 11 |
| SARS-CoV-2 | Blood and nose swabs | 36 | RNA-seq | 0, 0.25, 14, 28(blood) 0,1,3,5,7,10,14(nose swabs) | E-MTAB-12993 | 12 |
| H3N2 | Blood and nose swabs | 20 | RNA-seq | 0, 1, 2, 3, 7, 10, 14, and 28(blood) 0,1, 2,3,7,14(nose swabs) | EGAD50000000956 | |
| HRV | Nose swabs | 17 | Microarray (Affymetrix) | -14,0.33, 2.00 | GSE11348 | 13 |
| H1N1 | Whole blood | 21 | Microarray (Illumina) | 0, 1, 2, 3, 4 | GSE90732 | 14 |
| RSV | Nose swabs | 58 | RNA-seq | 0,3 | GSE155237 | 15 |
| H1N1 | PBMC | 24 | Microarray (Affymetrix) | 0, 0.21, 0.50, 0.90, 1.21, 1.50, 1.90, 2.21, 2.50, 2.90, 3.21, 3.50, 3.90, 4.21, 4.50 | GSE52428 | 16 |
| H3N2 | 17 |
We have collected transcriptomes from 18 cohorts across 10 inoculation studies, totaling 429 participants. This collection covers inoculations of five respiratory viruses: H1N1, H3N2, RSV, HRV, and SARS-CoV-2. The time span ranges from -14 to 28 days before and post for inoculation. The sample source includes blood (n=14) and nasal swabs (n=4). The transcriptomic data types include microarrays from Illumina (n=2) and Affy (n=11); bulk RNA-seq (n=5) ( Figure 4).
The complete list of vaccination datasets included in our collection can be found in Table 2.
| Pathogen | Vaccine type | Sample type | Sample size | Profiling platform | Time point | Accession number | Citation |
|---|---|---|---|---|---|---|---|
| Influenza | TIV | PBMC | 172 | Microarray (Illumina) | 0,2,7,28 | GSE107990 | 17 |
| Influenza | LAIV | Whole blood | 20 | Microarray (Illumina) | 0,1,7,30 | GSE52005 | 18 |
| Influenza | TIV | Whole blood | 17 | Microarray (Illumina) | 0,1,7,30 | ||
| Influenza | LAIV | PBMC | 28 | Microarray (Affymetrix) | 0,3,7 | GSE29619 | 19 |
| Influenza | TIV | PBMC | 28 | Microarray (Affymetrix) | 0,3,7 | ||
| Influenza | LAIV and inactivated | Blood | 44 | RNA-seq | 0,3,7,29,85,92,113 | GSE217770 | 20 |
| Influenza | inactivated | PBMC | 50 | RNA-seq | 0,1,3,7,21,22,24,28 | GSE102012 | 21 |
| Influenza | LAIV | Nasal epithelium | 40 | RNA-seq | 0,3 | GSE230494 | 22 |
| Influenza | LAIV | Nasal cells | 55 | RNA-seq | 0,5,12 | GSE117580 | 23 |
| Influenza | TIV | Nasal cells | 62 | RNA-seq | 0,5,12 | ||
| Influenza | vetor viral virus | Whole blood | 11 | Microarray (Illumina) | 0,0.5,1,2 | GSE61754 | 11 |
| Influenza | Inactivated | PBMC | 14 | RNA-seq | -7,0,1,2,3,4,5,6,7,8,9,10,21 | GSE45764 | 24 |
| Influenza | Inactivated | Whole blood | 18 | Microarray (Illumina) | -7,0,1,3,7,10,14,21,28 | GSE30101/GSE48762 | 25 |
| Influenza | Inactivated | PBMC | 212 | Microarray (Affymetrix) | 0,3,7,14 | GSE74817 | 26 |
| Influenza | Inactivated | Whole blood | 247 | Microarray (Illumina) | 0,1,3,14 | GSE48024 | |
| Influenza | Inactivated | Whole blood | 91 | Microarray (Illumina) | 0 | GSE41080 | |
| Influenza | Inactivated | PBMC | 5 | RNA-seq | 0,1,2,3,4,5,6,7,8,9,10 | GSE45735 | |
| Influenza | Inactivated | PBMC | 60 | Microarray (Illumina) | 0,2,4,7,28 | GSE59743/GSE95584 | |
| Influenza | Inactivated | PBMC | 27 | Microarray (Affymetrix) | 0,3,7 | GSE29617/GSE29614 | |
| Influenza | Inactivated | PBMC | 64 | Microarray (Illumina) | 0,2,4,7,28 | GSE59654 | |
| Influenza | Inactivated | Whole blood | 51 | Microarray (Illumina) | 0,2,7,28 | GSE101709 | |
| Influenza | Inactivated | PBMC | 42 | Microarray (Illumina) | 0,4,7,28 | GSE59635 | |
| Influenza | Inactivated | Whole blood | 44 | Microarray (Illumina) | 0,2,7,28 | GSE101710 | |
| Influenza | Inactivated | PBMC | 63 | Microarray (Affymetrix) | -7,0,1,7,70 | GSE47353 | |
| Influenza | LAIV | PBMC | 28 | Microarray (Affymetrix) | 0,3,7 | GSE29615 | |
| influenza | Inactivated | Whole blood | 123 | Microarray (Affymetrix) | −7,0,1,3,7,10,14,21,28 | SDY311/SDY312/SDY314/SDY315/SDY112 | 27 |
| Influenza | TIV | Whole blood | 34 | Microarray (Illumina) | 0,7,14 | SDY272/SDY648/SDY739/SDY819/SDY622 | 28 |
| influenza | TIV | Whole blood | 65 | RNA-seq | 0,2,7,28 | SDY1393 | 29 |
| Influenza | TIV | Whole blood | 6 | RNA-seq | 1,3,60 | SDY300 | * |
| COVID | mRNA | PBMC | 16 | scRNA | 0,7,21,28 | GSE247917 | 30 |
| COVID | mRNA | Blood | 23 | RNA-seq | 0,1,2,3,4,5,6,7,8,9 | GSE190001 | 31 |
| COVID | mRNA | PBMC | 214 | RNA-seq | 0,22,90,180,360 | GSE220682 | 32 |
| COVID | mRNA | Blood | 6 | scRNA | 0,1,2,7,21,22,28,42 | GSE171964 | 33 |
| COVID | mRNA | Blood | 56 | RNA-seq | 0,1,7,21,22,28 | GSE169159 | |
| COVID | mRNA | PBMC | 8 | scRNA | 28,35,60,110,201 | GSE195673 | 34 |
| COVID | mRNA | PBMC | 4 | scRNA | 0,7,30,90 | GSE210229 | 35 |
| COVID | vetor viral virus | Whole blood | 36 | RNA-seq | 0,50,57 | GSE228842 | 36 |
* This data is available at ImmPort (https://immport.org/shared/home) under study accession SDY300: Healthy Human DC and monocyte subsets transcriptional regulations in response to Fluzone 2010-2011 and pneumococcal vaccinations.
We have gathered transcriptomes from 37 cohorts across 22 vaccine studies, totaling 2084 participants. This collection focuses on studying Influenza and COVID-19 vaccines. The time span ranges from -28 to 360 days. The data source includes blood (n=34) and nasal swabs (n=3). The transcriptomic data types include microarrays from Illumina (n=13) and Affy (n=7); bulk RNA-seq (n=13) and scRNA-seq (n=4) ( Figure 5). Vaccine types: TIV (n=22) and LAIV (n=6) were used for influenza vaccination, mRNA vaccine (n=7) was used for COVID-19 vaccination, and vector adenovirus (n=2) was used for both influenza and COVID-19, one of each.
All datasets in our curated collection are also accessible to the public on the NCBI GEO website at https://www.ncbi.nlm.nih.gov/gds/ ArrayExpress at https://www.ebi.ac.uk/biostudies/arrayexpress/studies ImmPort at https://www.immport.org/shared/home, and the European Genome-phenome Archive (EGA)37 at https://ega-archive.org/datasets/ cited in the manuscript using its GEO number, ArrayExpress number, ImmPort study number or EGA study number. All these citation and accession numbers are listed in Table 1. The dataset can be found by searching for the accession number on the corresponding websites listed above. Note that downloading from ImmPort requires registering a personal account on their website.
NCBI: Expression Omnibus (GEO): Transcriptome datasets of the transcriptional response in blood samples following HRV, RSV, H3N2 and H1N1 inoculation. Accession Number: GSE73072; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE730728
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets of the transcriptional response in blood samples following HRV, RSV and H3N2 inoculation. Accession Number: GSE17156; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171569
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets of the transcriptional response in blood samples following H3N2 inoculation. Accession Number: GSE30550; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3055010
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets of the transcriptional response in blood samples following H3N2 inoculation. Accession Number: GSE61754; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE6175411
EMBL-EBI: ArrayExpress: Transcriptome datasets of the transcriptional response in blood and nasal samples following SARS-CoV-2 inoculation. Accession Number: E-MTAB-12993; https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-1299312
European Genome-Phenome Archive (EGA): Transcriptome datasets to study the transcriptional response in blood samples following H3N2 inoculation. Accession Number: EGAD50000000956; https://ega-archive.org/datasets/EGAD5000000095612 (Data available on request)
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in nasal samples following HRV inoculation. Accession Number: GSE11348; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1134814
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following H1N1 inoculation. Accession Number: GSE90732; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE9073215
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in nasal samples following RSV inoculation. Accession Number: GSE155237; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15523715
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following H1N1 and H3N2 inoculation. Accession Number: GSE52428; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5242816
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE107990; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE10799017
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE52005; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5200518
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE29619; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2961919
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE217770; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21777020
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE102012; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE10201221
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in nasal samples following influenza vaccination. Accession Number: GSE230494; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23049422
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in nasal samples following influenza vaccination. Accession Number: GSE117580; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE11758023
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE61754; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE6175411
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE45764; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4576424
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE30101 and GSE48762; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30101 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4876225
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE74817; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE7481726
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE48024; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4802426
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE41080; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4108026
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE45735; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4573526
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE59743 and GSE95584; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE59743 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE9558426
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE29617 and GSE29614; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29617 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2961426
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE59654; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5965426
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE101709; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE10170926
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE59635; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5963526
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE101710; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE10171026
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE47353; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4735326
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: GSE29615; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2961526
ImmPort: Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: SDY311, SDY312, SDY314, SDY315 and SDY112; https://www.immport.org/shared/study/SDY311, https://www.immport.org/shared/study/SDY312, https://www.immport.org/shared/study/SDY314, https://www.immport.org/shared/study/SDY315 and https://www.immport.org/shared/study/SDY11227
ImmPort: Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: SDY272, SDY648, SDY739, SDY819 and SDY622; https://www.immport.org/shared/study/SDY272, https://www.immport.org/shared/study/SDY648, https://www.immport.org/shared/study/SDY739, https://www.immport.org/shared/study/SDY819 and https://www.immport.org/shared/study/SDY62228
ImmPort: Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: SDY1393; https://www.immport.org/shared/study/SDY139329
ImmPort: Transcriptome datasets to study the transcriptional response in blood samples following influenza vaccination. Accession Number: SDY300; https://www.immport.org/shared/study/SDY300
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following COVID vaccination. Accession Number: GSE247917; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE24791730
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following COVID vaccination. Accession Number: GSE190001; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19000131
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following COVID vaccination. Accession Number: GSE220682; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22068232
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following COVID vaccination. Accession Number: GSE171964 and GSE169159; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE171964 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16915933
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following COVID vaccination. Accession Number: GSE195673; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19567334
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following COVID vaccination. Accession Number: GSE210229; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21022935
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in blood samples following COVID vaccination. Accession Number: GSE228842; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22884236
NCBI: Gene Expression Omnibus (GEO): Transcriptome datasets to study the transcriptional response in nasal samples following HRV inoculation. Accession Number: GSE11348; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1134813
We extend our gratitude to all the researchers who have publicly shared their datasets by depositing them in GEO, ArrayExpress, ImmPort and EGA.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Systems immunology, transcriptomics, metabolomics applied to the study of infectious and parasitic diseases.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |
|---|---|
| 1 | |
|
Version 1 13 May 25 |
read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)