Keywords
biocuration, information retreieval, text mining
This article is included in the EMBL-EBI collection.
This article is included in the AIDR: Artificial Intelligence for Data Discovery and Reuse collection.
Biomedical databases are an important part of the scientific infrastructure for organising and synergising research outputs. Many of these databases abstract content from the rapidly expanding scientific literature. Therefore, database curators require effective literature search methods in order to capture research relevant to their domain.
This article describes LitSieve, a literature search tool with filtering based on text mined annotations, and flexible article organisation features. It allows users to define filters based on biomedical entities like genes, diseases and species to include or exclude particular articles within their results. By combining a search query with a filter, curators are able to identify articles relevant to the database which they are curating. LitSieve uses APIs provided by Europe PMC, from which abstracts, article full text and text mined annotations are drawn.
LitSieve is available at https://www.ebi.ac.uk/europepmc/litsieve/
biocuration, information retreieval, text mining
Biomedical databases have become a critical infrastructure supporting life science research. Biologists and bioinformaticians depend on databases to interpret their results.1,2 Many important databases depend upon curation of the scientific literature in order to identify and extract relevant information into a structured format. When curating the literature, domain expert biocurators search and sort through scientific articles, and read those that appear relevant to their databases, focussing on the specific facts that they wish to capture.1 For example, a reference to a particular pair of proteins interacting, or an association between a gene and a disease.
Biocurators may use biomedical literature databases to identify ‘curatable’ literature. In this work we describe LitSieve, a system building on the Europe PMC database to provide literature filtering and organisational functions designed to assist with biocuration workflows.3 While there is a variety of literature search and organisation software already available, LitSieve provides a unique ability to filter based on a wide range of text-mined annotations.
Europe PMC is a comprehensive database of life science literature. It contains abstracts from PubMed and Agricola, full text articles from PubMed Central and content from 35 life science relevant preprint servers including bioRxiv and medRxiv. The database contains a total of over 45 million articles, and the full text of the article is searchable for 10 million of those. Literature in Europe PMC is enriched by over 2 billion text-mined annotations. Annotations are references to biomedical entities or concepts such as gene/protein names and diseases, extracted from the literature using a variety of methods. In total there are 43 different categories of annotation. These entities are normalised to an entry in a database. For example, species names are normalised to the NCBI taxonomy. These annotations are made available via a public REST API.3
A number of tools to assist biocurators have been developed. PubTator permits users to search based on six types of text mined ‘bioentities’–genes, diseases, chemicals, single nucleotide polymorphisms (SNPs), species and cell lines–against the PubMed and PubMed Central databases.4 Users are further able to search based on 12 types of text mined interactions between bio entities; for example drug interactions between two chemicals or causation between a SNP and a disease. It also allows users to gather articles into user defined ‘collections’.
LitSuggest uses machine learning (ML) to suggest similar articles to those selected by the user.5 Articles identified by the trained model can then be marked as relevant or irrelevant to further refine the model. In the context of curation, this permits biocurators to submit a list of articles they have already curated and ideally find further ‘curatable’ articles.
Tools using large language models (LLMs) to search and summarise literature have also emerged,6 however, these tools have yet to be comprehensively assessed in the context of biocuration. Given the impact of LLMs like ChatGPT on the wider technology landscape it seems inevitable that biocurators will use LLM based tools. However, their propensity for factual errors remains an open problem, and presents a challenge to deploying them on biocuration tasks, where statements must be reliably attributed.7
Development of LitSieve began with the goal of providing an interface to Europe PMC with improved utility for biocurators. The initial concept being that curators may prefer not to use certain ML-based suggestion or recommendation systems, due to their ‘black box’ nature.8 An internal survey of biocurators was conducted to understand their usage of literature search tools and the types of literature they were interested in. Possible features were discussed, and curators were observed while completing tasks. As development progressed, feedback from biocurators was incorporated into the prerelease versions at each stage.
The LitSieve system is based upon retrieval using a user-specified search query, the results of which are filtered as chosen by the user. This concept prioritises the explainability of the results, since it is clear to users exactly why a particular article has been included or excluded from their search results: Only search results which are retrieved by their boolean search term are included and, of those, only those that match all the filters are displayed in the search results. Therefore, the reason for the inclusion or exclusion of a particular article is always transparent (see Figure 1c for an illustration of a matching search result).
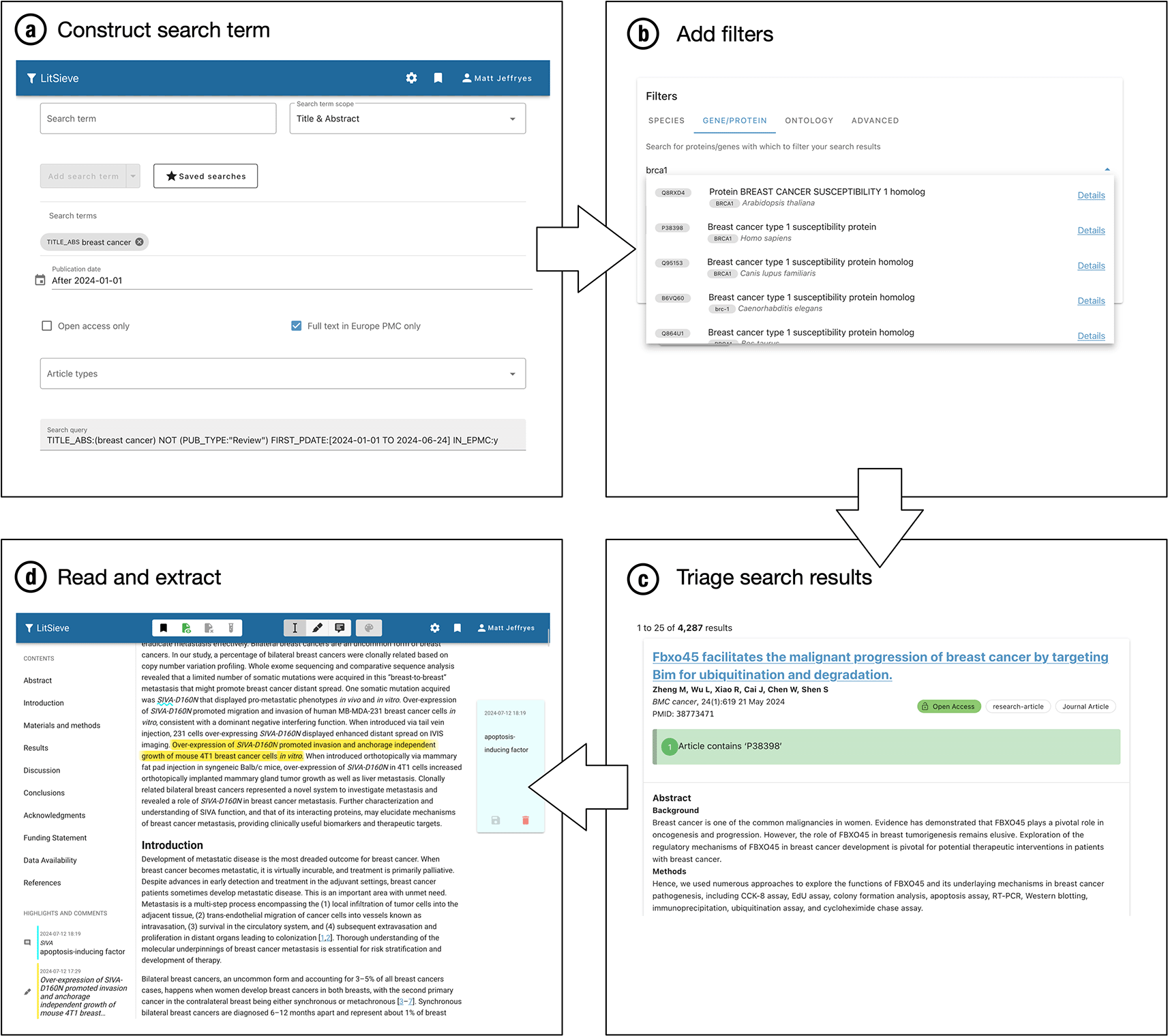
A literature search is performed (a), the results optionally filtered (b), and then the literature retrieved (c) which can be read and annotated (d) according to the requirements of the user.
Although ML is used to identify many of the text mined annotations used to filter, this approach reduces the scope of the ‘black box’ area of the retrieval system, it is smaller and more comprehensible. This modular, filter-based concept also enables additional filters to be developed and added, fitting within the same architecture.
LitSieve is a literature search and organisation tool designed for biocuration. It permits users to perform a standard literature search and then filter it based upon text-mined annotations. The filtering system is very flexible and accommodates a wide variety of use cases. An overview of the process of using LitSieve is shown in Figure 1.
LitSieve builds upon Europe PMC’s public articles and annotations APIs and is implemented using the Vue JavaScript framework. Searches are configured using a form and user selected parameters define which relevant articles are fetched from Europe PMC. These results may then be filtered according to the text-mined annotations found in the articles. Any of the 43 categories of annotation in Europe PMC can be used for filtering. Users can filter to include articles according to the presence or absence of a specific annotation. Three types of filter are available (include, exclude, ignore), listed in Table 1 and illustrated in Figure 2. Annotations are fetched from Europe PMC, and then used to filter the articles client-side. The basic search, filtering and reading functions can be used by anonymous users. Saving lists, highlights and notes requires users to register with either an email address or by using ORCID login.
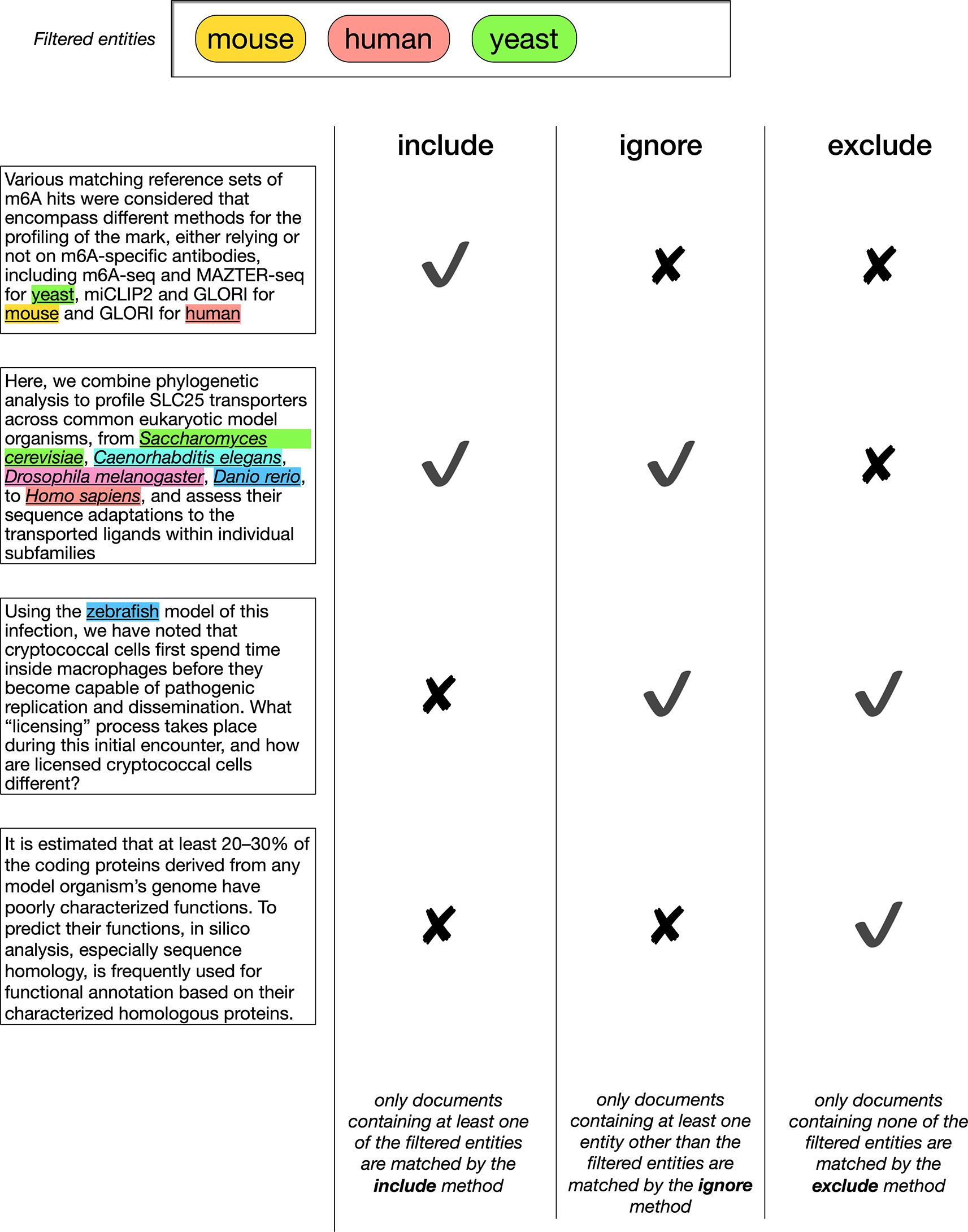
Three taxa are specified for filtering at the top. In the left column, 4 documents are shown. In reality, the filter would be applied using the entire document, or a specified section, but in this case a short fragment is used for illustrative purposes. In the fragments, all mentions of a species are underlined, and the specified species are highlighted. In the top row, each filter type is listed. Below the filter types it is indicated whether a filter of the corresponding type, with the three specified taxa, would result in the document being included in the search results or not.
The filter may be restricted to a specific section of the article (for example, finding only articles that have a ‘mouse’ annotation within their Methods section). Lists of identifiers may be saved for convenience, for example, if a curator has a list of diseases of interest that they wish to use as a filter on many searches.
For convenience, several types of annotation can be filtered using an integrated auto-complete interface. Species and other taxonomic ranks can be retrieved from the NCBI taxonomy [1], gene and protein names can be retrieved from UniProt, and terms from the Gene Ontology, Uberon, Experimental Factors Ontology, and Chebi can be retrieved from the Ontology Lookup Service.9,10
Articles found using LitSieve can be saved to lists. This accommodates a triage workflow where users can flag literature as either curatable or non-curatable, but users may organise lists as they wish, and no specific workflow is imposed. Articles may be added or removed from lists directly from the search result page, or from the reading view. This permits, for example, a curator to remove an article from their ‘triage’ list after having read it and found it to be non-curateable. The “quick lists” feature allows users to assign an icon and colour to a particular list, which permits easy visual identification of list membership in the search results page. This allows a curator to identify, for example, articles they have already triaged.
In the reader view, users may highlight and add private notes to articles (see Figure 1d). Biocurators may use this to highlight curatable passages from the article or other pertinent details such as cell lines used in experiments.
Users may recall and reorder saved articles from a list management view. A list of all articles to which notes or highlights have been added is also available. Lists may be used to organise or prioritise articles for curation, or to save a group of related articles.
IntAct is a molecular interaction database.11 It is essentially a graph of interacting molecules, with the vertices being biologically active molecules like proteins, and the edges denoting some kind of interaction between a pair of them. IntAct is manually curated; every interaction has been captured by a biocurator. This is a time intensive process, and given the available resources, prioritisation is necessary because not every possible interaction published can be incorporated into the database. As a strategic goal, IntAct has prioritised adding new molecules to the database (increasing the number of vertices) over adding edges between molecules already in the database, prioritising coverage over increasing the number of evidences for known relationships. Therefore, it is desirable to find literature that discusses protein–protein interactions where at least one of the proteins is not yet listed in IntAct.
LitSieve enables the IntAct biocurators to filter out articles that will not add new molecules to the database. After performing a literature search, an ‘ignore’ filter that lists UniProt identifiers for all proteins already present in IntAct can be applied. This will filter out any article that does NOT mention at least one protein not in the list specified by the user. That is, only articles mentioning proteins new to IntAct will be shown in the result list. While this does not guarantee that the article will discuss a curatable protein interaction, it will filter out articles which certainly do not increase the number of proteins covered by IntAct. In this way, LitSieve enables IntAct curators to perform literature searches constructed using their experience while benefiting from the text-mined annotations in Europe PMC to speed up their triage of the results. A step by step illustration of this workflow is available at 10.5281/zenodo.15682791.
UniProt is a data resource for protein sequence and functional information. One component of UniProt is the SwissProt subset of the UniProt knowledgebase (UniProtKB/SwissProt). This is a curated resource summarising experimental and computationally predicted functional information selected and reviewed by an expert biocurator. In order to carry out this work, UniProt biocurators search for, and read, literature related to the proteins which they are tasked with creating and updating records for.
LitSieve has been used to curate proteins related to antimicrobial resistance into UniProt. The ability to filter search results based on species is beneficial during triage to sift out articles not related to the entry being curated. Since a single species may be referred to by multiple different names (for example, mouse, mice, M. musculus, Mus musculus), filtering based on concept rather than exact text matches can save time and effort during the triage process.
LitSieve allows biocurators to combine their literature search expertise with filters based on text-mined annotations. This transparent and reproducible approach to literature discovery allows biocurators and other users to understand why a particular article has or has not been captured by their query. The flexible filter architecture permits use cases that we have not yet anticipated. Based on Europe PMC, LitSieve benefits from daily literature updates and can search across over 31 million abstracts and over 10 million full text articles. Filtering can be performed using 2 billion text-mined annotations in 43 categories. There are a variety of other tools available for biocuration literature search, however, to our knowledge, no others are able to search based on this number of types of annotation.
LitSieve provides an integrated interface for organising and prioritising literature. We anticipate that by integrating biocuration related features into a single application, biocuration workflows can be made more efficient.
LitSieve is available publicly at https://www.ebi.ac.uk/europepmc/litsieve/.
Source code is available in two repositories under an MIT licence. The front-end is available at https://gitlab.ebi.ac.uk/mjj/biocuration-toolbox , and the back-end is available at https://gitlab.ebi.ac.uk/mjj/litsieve-backend . An archived copy of these repositories at time of submission has been deposited in Zenodo: https://dx.doi.org/10.5281/zenodo.15480211.
All the authors contributed to conceptualisation and determining the methodology. HH, MH and JM provided supervision. MJ was responsible for software development, and for drafting the original manuscript. All authors contributed to review and editing.
We thank Islam Hassan, Mohamed Selim, and Jagadeeswararao Poluru for software engineering and Kalpana Panneerselvam, Paul Denny and other users for testing, and feedback. This work was supported by the European Molecular Biology Laboratory (EMBL).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Software engineering. Bioinformatics.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |
|---|---|
| 1 | |
|
Version 1 11 Jul 25 |
read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)