Keywords
Molecular mechanisms, Drug resistance, infectious diseases, public health & interventions
This article is included in the Antimicrobial Resistance collection.
Drug resistance in infectious diseases present a major global health concern, reducing treatment efficacy and increasing morbidity and mortality. Resistance arises from different molecular mechanisms including genetic mutations, enzymatic degradation of drugs, alterations in target sites, efflux pump overexpression, and reduced membrane permeability. These mechanisms contribute to the development of multidrug-resistant and extensively drug-resistant pathogens across bacterial, viral, and mycobacterial species. Scientist have developed therapeutic strategies to control these mechanisms of multidrug-resistant. These include the development of novel antimicrobials such as teixobactin and pretomanid, the application of β-lactamase inhibitors, rational drug combinations, host-directed therapies, and antimicrobial peptides. Advances in biotechnology have enabled precise-targeted approaches, including phage therapy, CRISPR-based antimicrobials, and nanocarrier-mediated drug delivery. Public health interventions are important in reducing the burden of AMR. These include global surveillance systems, antimicrobial stewardship programs, vaccination, infection control protocols, and regulatory policies governing antimicrobial use in humans and animals. Future directions emphasize the integration of precision medicine, artificial intelligence, environmental monitoring, and international governance to strengthen AMR control. A sustained and coordinated global response is essential to preserve the efficacy of current therapies and promote the development of new interventions.
Molecular mechanisms, Drug resistance, infectious diseases, public health & interventions
Drug resistance in infectious diseases present a global health challenge, posing a significant threat to modern medicine and public health. It occurs when pathogens, including bacteria, viruses, fungi, and parasites developed defensive mechanisms to survive the exposure of antimicrobial agents. The efficacy of the drug is compromised by this process resulting to persistent infections, higher mortality rates, and the widespread transmission of resistant microorganisms.1 The Lancet’s global burden study on bacterial antimicrobial resistance (1990–2021) reported that AMR was directly responsible for an estimated 1.27 million deaths and contributed to nearly 5 million deaths worldwide, surpassing mortality rates from HIV/AIDS and malaria.2 This burden has been reported to be pronounce in low- and middle-income countries (LMICs), where weak surveillance systems, unregulated drug use, and limited access to second-line therapies amplify the spread the of resistant infections.3 Extensively drug-resistant (XDR) and multidrug-resistant (MDR) bacteria strains including drug-resistant HIV, multidrug-resistant Mycobacterium tuberculosis (MDR-TB), and methicillin-resistant Staphylococcus aureus (MRSA) has complicate clinical management.4 Historically, the development of antibiotics and antiviral drugs has transformed the treatment of infectious diseases. However, the growing incidence of antimicrobial resistance (AMR) as a result of factors such as antibiotic misuse, overuse, inappropriate prescribing practices, and inadequate infection control measures. These factors have promoted antimicrobial resistance (AMR) more than the rate at which new drugs are being discovered placing a severe constraint on healthcare system. Drug-resistant infections are currently considered to be one of the biggest threats to global health, contributing estimated 700,000 deaths annually.5 Treatment for resistant infections is often prolong leading to increase healthcare cost and risk of treatment-related complications. This results to reduces workforce productivity and places immense strain on health systems particularly in low- and middle-income.6 Addressing AMR requires a comprehensive and coordinated approach that integrates molecular research, innovative pharmacological strategies, strengthened public health policies, and global collaboration.
The molecular mechanisms of bacterial antibiotic resistance involve several key processes, including genetic mutations, enzymatic modification or degradation of antibiotics, alteration of drug targets, and the overexpression of drug efflux pumps as depicted by Figure 1. Antimicrobial resistance (AMR) exhibits pathogen-specific molecular profiles and epidemiological behaviors that require differentiated surveillance and treatment. Table 1 summarizes key pathogen-specific resistance mechanisms, their clinical and public health implications, available treatment options.
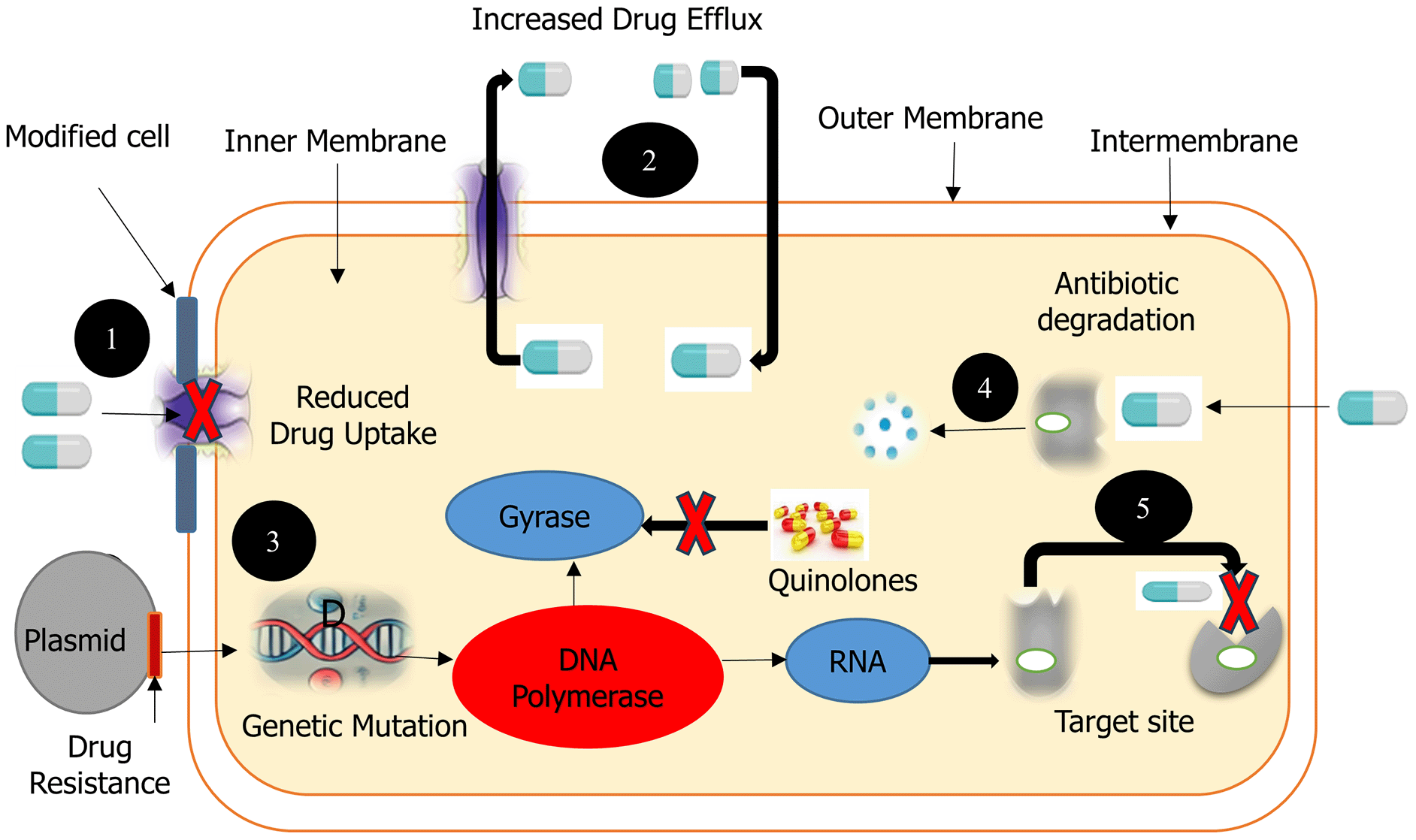
This diagram illustrates the key strategies employed by bacteria to resist the effects of antibiotics. These mechanisms include: (1) Reduced Drug Uptake: Alterations in the bacterial cell envelope, such as modifications to porin channels, limit the entry of antibiotics. (2) Efflux Pumps: Transmembrane protein pumps actively export antibiotics from the bacterial cytoplasm, reducing intracellular drug concentrations. (3) Genetic Mutation and Target Modification: Mutations in chromosomal genes (e.g., those encoding DNA Polymerases or Gyrase, as shown with quinolones) or acquisition of resistance genes via plasmids can alter the antibiotic's molecular target, preventing effective binding. (4) Antibiotic Degradation: Enzymes produced by the bacterium chemically modify or degrade the antibiotic molecule, rendering it inactive. (5) Target Site Modification: Structural changes at the antibiotic's binding site within the bacterial cell prevent the drug from exerting its inhibitory effect, even if it enters the cell.
Efflux pumps can be categorized into multiple groups according on their energy source and structure. Major Facilitator Superfamily (MFS) transporters and ATP-Binding Cassette (ABC) transporters are two of the most significant families of efflux pumps frequently linked to drug resistance. Drugs and other substrates are actively transported across the cell membrane by ABC transporters, which use the energy produced by the hydrolysis of ATP. These pumps can expel variety of antibiotics and antibacterial compounds due to their broad substrate specificity.7 P-glycoproteins (P-gp), members of ATP-binding cassette (ABC) transporter family involved in drug resistance and frequently present in both cancer cells and microorganisms. These transmembrane proteins utilize ATP hydrolysis to actively transport a wide range of substrates including chemotherapeutic agents and antibiotics out of cells, thereby reducing intracellular drug concentrations and reducing therapeutic efficacy. ABC transporters have been linked to resistance in bacterial infections to antibiotics like macrolides, fluoroquinolones, and aminoglycosides.8 Major Facilitator Superfamily (MFS) transporters are secondary active transport proteins that contribute significantly to antibiotic resistance by expelling drugs from bacterial cells. The MFS transporters make use of ion gradients across the membrane or the proton motive force (PMF) to expel drugs out of bacterial cells. Both Gram-positive and Gram-negative bacteria exhibit MFS transporters. The NorA efflux pump is another MFS transporter which helps Staphylococcus aureus to develop quinolone resistance by removing fluoroquinolones out of the cell. MFS transporters can also pump several antibiotic classes, such as β-lactams, chloramphenicol, and tetracyclines out of the cell rendering them less effective.9 The expression of these efflux pump can be upregulated by pathogens in reaction to antibiotics or other antimicrobial treatments. Many bacterial species, such as Pseudomonas aeruginosa and Escherichia coli have been reported to overexpress efflux pumps. In these organisms, the MexAB-OprM efflux system plays a role in resistance against β-lactams, aminoglycosides, and fluoroquinolones. Efflux pumps frequently function in synergy with other resistance mechanisms, like target site alteration or enzymatic drug degradation.10 Recent evidence suggests that efflux pump expression can be regulated by environmental stress and antibiotic exposure. For example, E. coli upregulates AcrAB-TolC in response to sub-inhibitory concentrations of antibiotics, contributing to treatment failure and chronic infections. Chemotherapeutic drugs can also be expelled from tumor cells through efflux pumps during cancer treatment, which could result in multidrug resistance and a relapse of the disease. Treatment failures have been reported in diseases like tuberculosis, malaria, and other bacterial infections mediated by bacterial efflux pump.11
Genetic mutations can alter the genetic composition of bacteria, viruses, and other microbes, enabling them to survive treatments that would normally kill them or suppress their growth. These mutations can affect the structure of drug targets, reduce drug efficacy, or enhance mechanisms that allow microbes to withstand treatment.12 A point mutation involves a change in a single nucleotide base in an organism’s DNA sequence. This minor alteration can affect the functionality of proteins that serves as drug target receptors. These mutations cause changes in the structure of the RNA polymerase enzyme, reducing the drug’s ability to bind to and inhibit its activity.13 Point mutations can contribute to resistance in viral infections such as HIV resistance to antiretroviral drugs. Mutations in the reverse transcriptase gene, such as K103N, can lead to resistance against non-nucleoside reverse transcriptase inhibitors (NNRTIs) like efavirenz and nevirapine.14 Genetic mutations can also occur in the form of insertions or deletions of small nucleotide regions within the microbial genome. These can lead to resistance by either creating novel pathways or altering the functionality of drug targets. As a result, the drug becomes less effective because insertions or deletions in the bacterial genome prevent the drug from binding effectively to its target sites.15 For example, insertions in the gyrA and parC genes, which encode parts of DNA gyrase and topoisomerase IV, contribute to quinolone resistance in Escherichia coli and other Gram-negative bacteria. These mutations prevent quinolones from binding to these enzymes which are essential for bacterial DNA replication.16 Gene duplications or amplifications are another form of genetic mutation that can lead to drug resistance. This process involves the overproduction of resistance enzymes or drug targets, which can overwhelm the therapeutic efficacy of drugs. In cancer cells, multidrug resistance is often associated with increased synthesis of P-glycoprotein, a drug efflux pump that actively transports chemotherapeutic drugs out of the cells.17 This overproduction is caused by amplification of the MDR1 gene resulting to reduced drug accumulation and resistance. A similar mechanism is observed in microbial drug resistance, where the overproduction of a target enzyme can dilute the drug’s potency, commonly seen in cancer cells. In the context of infectious diseases, genetic mutations in drug targets are a primary cause of resistance to first-line drugs, such as those used to treat tuberculosis (TB). Isoniazid resistance is linked to mutations in the katG gene, which reduce the enzyme’s ability to activate the drug within the bacterial cell. Similarly, resistance to rifampicin arises from mutations in the rpoB gene, which codes for the beta subunit of RNA polymerase, disrupting the drug’s binding and effectiveness.18
The beta-lactam ring, present in antibiotics including cephalosporins, carbapenems, and penicillins are susceptible to bacterial enzymes beta-lactamases. These enzymes hydrolyze the β-lactam ring and deactivate the antimicrobial agents preventing them from inhibiting bacterial cell synthesis.19 β-lactamases are broadly classified into four molecular classes (A–D) based on their amino acid sequences and functional characteristics. Class A enzymes include extended-spectrum β-lactamases (ESBLs), which hydrolyze a wide range of cephalosporins and are commonly found in Escherichia coli and Klebsiella pneumoniae. Class B enzymes are metallo-β-lactamases (MBLs) that require zinc ions for activity and are capable of degrading carbapenems. Class C such as AmpC β-lactamases, chromosomally encoded in Enterobacter species can develop resistance to cephamycins and are not inhibited by clavulanic acid. Recent systematic review and Mata-analysis reported that 20.76% of E. coli isolates in Sub-Saharan Africa now express ESBLs, highlighting the urgent need for novel β-lactamase inhibitors such as avibactam and relebactam.20 Class D enzymes include oxacillinases (OXA-type), which are associated with resistance in Acinetobacter baumannii and other Gram-negative pathogens. The spread of β-lactamase genes frequently through plasmids and other mobile genetic elements has led to widespread multidrug resistance, particularly in nosocomial infections. The clinical implications are severe, as treatment options become limited leading to increased morbidity and higher mortality rates.21 Furthermore, Aminoglycoside-modifying enzymes, such as adenyl transferases, phosphotransferases, and acetyltransferases, are produced by certain pathogens to modify aminoglycosides (e.g., streptomycin and gentamicin). These modifications include adenylation, phosphorylation, or acetylation, rendering the antibiotics inactive by preventing them from binding to bacterial ribosomes. For example, in Pseudomonas aeruginosa and Staphylococcus aureus, the modification of these enzymes contributes to resistance against aminoglycosides, preventing antibiotics from interfering with bacterial protein synthesis.22 Similarly, bacterial resistance to chloramphenicol and macrolides (e.g., erythromycin) is mediated by enzymes that inactivate these drugs. Esterase and chloramphenicol acetyltransferases acetylate or hydrolyze macrolides and chloramphenicol, respectively, preventing them from binding to bacterial ribosomes. As a result, the antibiotics lose their ability to inhibit protein synthesis, which is their primary mechanism of action.23
Numerous antibiotics such as aminoglycosides, tetracycline, and macrolides interfere with the bacterial ribosome to inhibit their protein synthesis.24 The structure of the ribosome is altered by mutations in the rRNA-encoding genes, especially in the 23S and 16S rRNA subunits, which inhibits the efficient binding of antibiotics. Alterations in the 23S rRNA gene result in resistance to macrolides, such erythromycin, in Streptococcus pneumoniae. These alterations decrease drug’s affinity for binding to the ribosome sub-units. Penicillin and cephalosporin are examples of beta-lactam antibiotics that target Penicillin-Binding Proteins (PBPs) involved in the formation of bacterial cell walls. Resistance results from PBP mutations or structural changes that reduced the drug’s capacity to bind to the enzyme. For example, the mecA gene in Methicillin-resistant Staphylococcus aureus (MRSA) produces a modified PBP called PBP2a, which has a poor affinity for beta-lactam antibiotics.25 Fluoroquinolones like ciprofloxacin work by targeting DNA gyrase and topoisomerase IV to prevent the replication of bacterial DNA. Fluoroquinolone resistance is caused by mutations in the gyrA and parC genes, which encode these enzymes and decrease drug binding. For example, decreased susceptibility to fluoroquinolones due to mutations in DNA gyrase is common in bacteria species like Salmonella spp. and Escherichia coli.26 Viral enzymes like proteases required for viral replication are majorly targeted by antiviral drugs. Mutations in these enzymes can change the drug-binding site resulting to a resistance specie. For example, mutations in the HIV-1 protease gene result in resistance to protease inhibitors (lopinavir and ritonavir).27 These mutations alter the structure of the enzyme, reducing the effectiveness of drugs such as nucleoside reverse transcriptase inhibitors (NRTIs) and non-nucleoside reverse transcriptase inhibitors (NNRTIs).28 Mutations like K103N in the reverse transcriptase gene have been reported to be associated with resistance to NNRTIs such as efavirenz and nevirapine. These alterations prevent the inhibitors from binding to the reverse transcriptase enzyme, allowing the virus to continue replicating despite the presence of the drug. Additionally, antibiotic resistance in tuberculosis particularly involving first-line drugs such as isoniazid and rifampin. Resistance to isoniazid have been reported to be associated with mutations in the katG gene encoding for catalase-peroxidase enzyme essential for drug activation in Mycobacterium tuberculosis. Likewise, mutations in the rpoB gene responsible for encoding the β-subunit of RNA polymerase led to rifampin resistance by altering the drug’s binding site. Both mechanisms have been reported to compromised the treatment of Tuberculosis thus making it more difficult to cure.29
One of the most promising discoveries in recent years is teixobactin, a compound that targets lipid II and lipid III, essential precursors in bacterial cell wall synthesis. Unlike traditional antibiotics, teixobactin binds to highly conserved, non-protein cellular components, making it less susceptible to resistance development. It is efficacy against multidrug-resistant Gram-positive bacteria such as Staphylococcus aureus and Enterococcus faecalis.30 Another innovation is lefamulin, a pleuromutilin antibiotic that has been approved to treat community-acquired bacterial pneumonia. Lefamulin inhibits bacterial protein synthesis by binding to the peptidyl transferase center of the 50S ribosomal subunit, which is unique from other ribosome-targeting drugs. Its broad-spectrum action, oral bioavailability, and minimal resistance potential make it appropriate for respiratory tract infections, especially those caused by multidrug-resistant pathogens.31 The use of pretomanid in conjunction with bedaquiline and linezolid to treat MDR and XDR-TB has demonstrated sterilizing and bactericidal effects. Pretomanid inhibits mycolic acid production and produces reactive nitrogen species under anaerobic circumstances, resulting in significant antimycobacterial action.32
Agents such as avibactam, relebactam, and vaborbactam exhibit broad-spectrum inhibitory activity against class A and class C β-lactamases, including carbapenemases like KPC. These compounds are often co-administered with partner β-lactams such as ceftazidime, imipenem, and meropenem to restore their antimicrobial activity.33 Unlike traditional inhibitors such as clavulanic acid, which have limited efficacy against newer β-lactamases, these novel agents offer enhanced binding affinity and resistance to hydrolysis. This class of therapy not only expands treatment options for infections caused by extended-spectrum β-lactamase (ESBL) and carbapenem-resistant. Enterobacteriaceae (CRE), but also reduces the need for last-resort antibiotics like colistin.34
Combination regimens reduce the likelihood of resistance development and often result in synergistic antimicrobial effects. The most successful example of this approach is Highly Active Antiretroviral Therapy (HAART) for HIV/AIDS. The combinations of nucleoside reverse transcriptase inhibitors, non-nucleoside reverse transcriptase inhibitors, and protease inhibitors are used to suppress viral replication and prevent drug resistance.35 Similarly, the treatment of tuberculosis relies on the standard four-drug regimen comprising isoniazid, rifampin, ethambutol, and pyrazinamide. This multidrug approach prevents the selection of resistant subpopulations and achieves both sterilizing and preventive therapeutic goals. For multidrug-resistant TB, second-line regimens incorporating linezolid, bedaquiline, and delamanid have been developed, based on molecular susceptibility testing.36 In bacterial infections, combination therapies are used to improve efficacy in severe or polymicrobial infections. For example, the co-administration of aminoglycosides with β-lactams or glycopeptides enhances membrane penetration and protein synthesis inhibition.37
Host-directed therapies are used to treat diseases like tuberculosis, HIV, and sepsis, where excessive inflammation or immune dysfunction contributes significantly to pathology. The development of monoclonal antibodies has proven to be significant in host-directed therapy. For example, bezlotoxumab, an antibody targeting Clostridium difficile toxin B, reduces recurrence rates and severity of C. difficile infections when used alongside antibiotics.38 In viral diseases, neutralizing monoclonal antibodies have demonstrated efficacy against respiratory syncytial virus (RSV) and SARS-CoV-2, providing both prophylactic and therapeutic benefits. Furthermore, cytokine-based therapies and immune checkpoint modulators are under investigation for their ability to fine-tune the immune response in chronic infections and in immunocompromised individuals.39
Antimicrobial peptides (AMPs) are part of the innate immune system and serve as a frontline defense against pathogens by disrupting microbial membranes. Unlike traditional antibiotics, AMPs act through rapid, non-specific mechanisms that reduce the likelihood of resistance. Colistin, a polymyxin AMP, has been reintroduced as a last-resort agent for Gram-negative infections, especially those caused by carbapenem-resistant Acinetobacter and Klebsiella species.40 Furthermore, advancements in synthetic biology have led to the design of engineered bacteriocins and phage-derived lysins that selectively target pathogenic bacteria without affecting commensals. These biologics act by degrading bacterial cell walls or membranes, offering a pathogen-specific alternative to broad-spectrum antibiotics.41
Public health interventions are important in global response to antimicrobial resistance (AMR), complementing therapeutic innovations with system-wide strategies to prevent infection, monitor resistance trends, and promote responsible antimicrobial use.
Effective surveillance systems form the foundation for detecting, monitoring, and responding to emerging resistance trends. The WHO’s Global Antimicrobial Resistance Surveillance System (GLASS) was launched to standardize AMR data collection across human, animal, and environmental sectors. GLASS integrates microbiological, clinical, and antimicrobial consumption data to support evidence-based policy-making and early warning systems.42 This program is particularly important in Low- and middle-income countries (LMICs) which often lack laboratory infrastructure, trained personnel, and reporting frameworks. International initiatives such as the Fleming Fund and the Tripartite Alliance (WHO, FAO, WOAH) are supporting LMICs in building laboratory networks, improving diagnostics, and establishing national AMR action plans. In the United States, the National Healthcare Safety Network (NHSN) provides comprehensive resistance data across healthcare institutions, which informs antimicrobial stewardship and infection control practices.43
Antibiotic stewardship programs (ASPs) rely on multidisciplinary teams including infectious disease specialists, pharmacists, microbiologists, and epidemiologists. These multidisciplinary teams oversee prescribing practices and ensure that antimicrobials are used correctly. Key stewardship strategies include the selection of appropriate empirical therapies based on local resistance patterns, prompt de-escalation to narrow-spectrum agents once culture results are available, and defining optimal treatment durations. The Programs also incorporate prescriber education, audit and feedback mechanisms, and the integration of clinical decision support systems (CDSS).44 Evidence from high-income countries has shown that ASPs reduce antimicrobial use by up to 30%, decrease hospital-acquired infections such as Clostridium difficile, and lower resistance rates without compromising clinical outcomes. In the United Kingdom, the NHS National Stewardship Program successfully integrated stewardship into national policy, reducing unnecessary prescriptions and achieving notable reductions in MRSA and VRE incidence.45
Reducing the transmission of resistant pathogens is a public health is important, especially in hospitals where invasive procedures, immunosuppressed patients, and high antibiotic use. Standard infection prevention and control (IPC) measures include hand hygiene, contact precautions, environmental cleaning, and the use of personal protective equipment (PPE). These measures are particularly vital in intensive care units, long-term care facilities, and neonatal wards.46 Healthcare-associated infections caused by resistant organisms such as MRSA, carbapenem-resistant Enterobacteriaceae (CRE), and vancomycin-resistant Enterococcus (VRE) can be reduced by proper IPC practices. Novel technologies such as ultraviolet-C light disinfection and antimicrobial surface coatings are also being explored to reduce environmental contamination.47 In community settings, interventions focus on improving sanitation, access to clean water, and vaccination coverage to reduce the incidence of infectious diseases and, by extension, the need for antibiotics. For example, pneumococcal and Haemophilus influenzae type b (Hib) vaccines have contributed to declining antibiotic use by preventing bacterial pneumonia and meningitis.
Public education campaigns are important in reshaping health-seeking behavior and reducing antibiotics misuse. The WHO’s World Antimicrobial Awareness Week (WAAW) is observed globally to educate populations about the risks of antibiotic overuse and the importance of following prescription guidelines. Behavior changes communication strategies that involve healthcare providers, pharmacists, schools, and community leaders are more likely to succeed. For example, community-based education programs in Thailand and Vietnam have demonstrated measurable improvements in antibiotic awareness and reduced over-the-counter antibiotic sales.48
Strong regulatory frameworks are essential for limiting unregulated antimicrobial distribution, incentivizing responsible use, and supporting research and development. The European Union has set global standards by banning the use of antibiotics as growth promoters in livestock since 2006 and restricting prophylactic use.49 Global policy efforts have been catalyzed by the WHO’s Global Action Plan on Antimicrobial Resistance, which outlines five strategic objectives: improve awareness, strengthen surveillance, reduce infection incidence, optimize antimicrobial use, and foster sustainable investment in R&D. National action plans aligned with this framework are now in place in over 140 countries, though operationalization and financing remain key barriers.50 Innovative legal mechanisms like the U.S. GAIN Act (Generating Antibiotic Incentives Now) provide extended market exclusivity for companies developing drugs against priority pathogens. These incentives, alongside public-private partnerships, aim to revitalize antimicrobial pipelines that have been commercially neglected due to high development costs and low return on investment.51
The One Health approach recognizes the interconnectedness of these domains and promotes integrated interventions. Zoonotic transmission of resistant bacteria caused by antibiotic use in agriculture makes cross-sectoral strategies essential. Collaborative programs such as the Global Antibiotic Research and Development Partnership (GARDP) and ReAct facilitate global research, equitable access to antibiotics, and advocacy for sustainable use. Additionally, platforms like The Access to Medicine Index monitor pharmaceutical companies’ commitments to responsible antibiotic marketing and stewardship.52
Despite advances in antimicrobial discovery, surveillance, and public health intervention, antimicrobial resistance (AMR) continues to evolve and spread globally. The increasing complexity of resistance mechanisms and their cross-border implications demand innovative and forward-looking strategies.
The future of antimicrobial therapy lies in precision medicine customizing treatment based on individual patient profiles, pathogen genomics, and local resistance trends. Advances in pharmacogenomics, host-pathogen interaction studies, and rapid molecular diagnostics will allow clinicians to select optimal therapies with greater accuracy. For example, integration of whole-genome sequencing (WGS) with electronic health records will enable the identification of resistance genes in real time, guiding tailored treatment decisions while minimizing the use of broad-spectrum agents.53
Artificial intelligence (AI) and machine learning are increasingly being applied to AMR prediction, surveillance, and drug discovery. AI can analyze different datasets nincluding clinical records, genomic sequences, and global resistance patterns to identify emerging trends and forecast outbreaks. Tools such as DeepARG, ResFinder, and AMRPlusPlus use machine learning to detect known and novel resistance genes from metagenomic data. In drug development, AI enable the screening of chemical libraries, identifies synergistic drug combinations, and predicts molecular interactions with bacterial targets. These reduce development costs and shorten timelines, making antibiotic innovation more feasible. Additionally, AI-driven decision support systems in hospitals can provide real-time guidance on antimicrobial choice, reducing diagnostic uncertainty and empirical overuse.54
Bacteriophage engineering, antibody-antibiotic conjugates, and CRISPR-based antimicrobials offer pathogen-specific activity and reduced disruption to the microbiome. Engineered phages can be designed to target multidrug-resistant strains and even deliver CRISPR-Cas systems that selectively disrupt resistance genes within bacterial genomes. Researchers can use CRISPR to knock out specific resistance genes in bacteria, revealing their involvement in drug resistance and possibly reversing resistance phenotypes.55 The development of nanocarrier-based drug delivery systems, which enhance antimicrobial penetration, reduce toxicity, and circumvent efflux pumps. Liposomal formulations and polymer-based nanoparticles have demonstrated improved efficacy against resistant pathogens by targeting drug release at infection sites and evading degradation.56 For example, in animal models, lipid nanoparticles (LNPs) carrying CRISPR-Cas9 targeting the HIV long terminal repeat (LTR) regions to knock out the proviral DNA as depicted in Figure 2. Antibody-coupled Nanoparticles (LNPs) coupled with anti-CD4 antibodies has been used to delivered CRISPR-Cas9 to CD4+ T cells. Nanoparticles can administer CRISPR-Cas9 alongside immune-stimulating drugs or ARVs to eliminate latent virus while suppressing active replication. Gold nanoparticles (AuNPs) are usually functionalized with both CRISPR components and interleukin-15 (IL-15), resulting in viral DNA excision and increased immune activation.
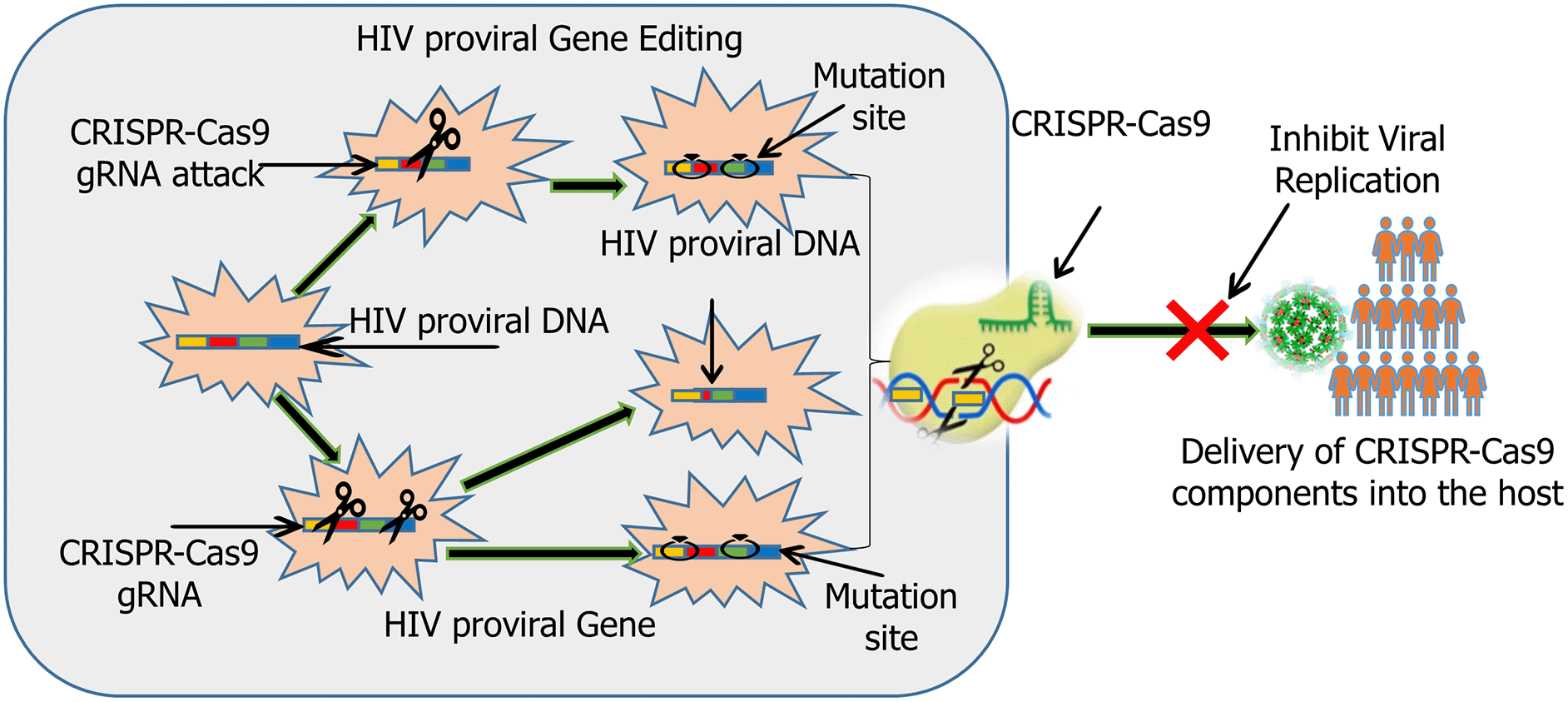
This diagram illustrates the mechanisms by which CRISPR-Cas9 technology is employed to combat HIV infection. CRISPR-Cas9, guided by specific guide RNAs (gRNAs), precisely targets the integrated HIV proviral DNA within the host cell genome. This targeting can lead to two primary outcomes: (1) HIV Proviral Gene Editing, where mutations are introduced at critical sites within the proviral DNA, rendering the virus replication-incompetent; or (2) HIV Proviral DNA Excision, leading to the complete removal of the integrated viral genome from the host cell. Both strategies aim to disrupt the viral life cycle by blocking the transcription and replication of new viral particles, ultimately preventing the spread of infection and the progression of HIV disease within an individual.
Addressing AMR from a One Health perspective requires environmental and agricultural interventions that limit the dissemination of resistance genes in natural ecosystems. Surveillance of antibiotic residues in water bodies, manure runoffs, and aquaculture systems is essential to understanding the environmental resistome.57
Future interventions should include:
○ Development of eco-friendly wastewater treatment technologies that remove antibiotic residues and resistant bacteria.
○ Implementation of antibiotic-free farming models, including alternatives such as probiotics, vaccines, and bacteriophage therapy in livestock.
○ Genetic monitoring of microbial populations in soil and water to detect the early emergence of resistance genes that may re-enter the clinical sphere via zoonotic transfer.
Investments in environmental biotechnology and ecological risk assessment frameworks will be crucial to mitigating resistance beyond clinical settings.
The sustainable control of AMR will require enhanced global governance mechanisms supported by long-term investment. Future frameworks should include binding international agreements on antibiotic use, equitable access to new drugs, and transparency in pharmaceutical R&D. A Global Treaty on Antimicrobial Use and Innovation, akin to climate change treaties, has been proposed by several health policy groups to coordinate cross-national action. Support for national AMR action plans, particularly in LMICs, will require integrated funding streams from international donors, development banks, and domestic governments.58
Future control of AMR will depend on sustained investment in basic and translational research, as well as the expansion of global collaborations. Research into resistance evolution, microbiome dynamics, and host-pathogen interactions will yield new targets and therapeutic strategies. Additionally, fostering a new generation of antimicrobial scientists through interdisciplinary training programs and international exchanges will build the global research capacity required to tackle AMR. Collaborative platforms like the International Centre for Antimicrobial Resistance Solutions (ICARS) and JPIAMR (Joint Programming Initiative on AMR) already support this vision by funding transnational research and innovation networks. Global data-sharing initiatives and open-access repositories will continue to be essential for equitable access to knowledge, diagnostics, and drug technologies.59
Antimicrobial resistance remains a major global health concern, driven by diverse molecular mechanisms that compromise the effectiveness of existing therapeutic agents. Resistance pathways such as enzymatic inactivation, efflux pump activity, target site alteration, and genetic mutations continue to evolve across a wide range of pathogens. These mechanisms contribute to treatment failures, increased disease burden, and growing healthcare challenges, particularly in resource-limited settings. Advances in therapeutic strategies, including the development of novel antimicrobials, β-lactamase inhibitors, and host-directed therapies, provide promising options for addressing resistant infections. Innovations in precision drug delivery, bacteriophage applications, and CRISPR-based technologies represent important future directions. Public health measures such as antimicrobial stewardship, surveillance systems, regulatory interventions, and infection prevention are essential for containing resistance and preserving therapeutic efficacy.
Future control efforts will require sustained investment in research, global collaboration, and integration of digital tools such as artificial intelligence and genomic surveillance. Strengthening cross-sectoral coordination through One Health frameworks and international policy alignment will be critical for effective and equitable responses. A multidisciplinary and systems-based approach is essential to limit the spread of antimicrobial resistance and safeguard public health.
All data supporting the findings of this study are contained within the article. Additional materials, including figures and extended datasets, have been deposited in a public repository and are openly accessible.60
Figshare: https://doi.org/10.6084/m9.figshare.29701106.v260
This project contains the following extended data:
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the topic of the review discussed comprehensively in the context of the current literature?
Partly
Are all factual statements correct and adequately supported by citations?
Yes
Is the review written in accessible language?
Yes
Are the conclusions drawn appropriate in the context of the current research literature?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Microbiology, AMR, Genomics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |
|---|---|
| 1 | |
|
Version 1 12 Aug 25 |
read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)