Keywords
Type 2 diabetes mellitus, Diabetic kidney disease, Metabolomic study, Biomarker identification, Pathway analysis.
Diabetic Kidney Disease (DKD) is one of the microvascular complications of Type 2 Diabetes Mellitus (T2DM) which tended not to be detected early, necessitating more effective biomarkers for its detection. HbA1c showed a negative correlation with the estimated glomerular filtration rate (eGFR) and was significantly associated with an increased urine albumin-creatinine ratio (UACR), thereby influencing the progression of DKD.
This study was an untargeted metabolomics study and aimed to compare the urinary metabolites of low DKD risk T2DM patients with controlled and uncontrolled HbA1c in patients taking metformin-glimepiride.
This study conducted as a cross-sectional study using non-probability sampling at Pasar Minggu District Health Center, 32 samples were divided into controlled (n=16) and uncontrolled HbA1c groups (n=16). Blood samples were collected for HbA1c and eGFR measurements, while urine samples were analyzed for UACR and metabolites. Analysis was performed using LC/MS-QTOF and the data was processed using MetaboAnalyst 6.0 and various databases. The discrimination results of the two groups were visualized through Principal Component Analysis (PCA) and Partial Least Squares-Discriminant Analysis (PLS-DA). The significance of metabolites between groups was selected with the parameters VIP>1, log2(FC)>1.2, and p-value<0.05.
Fifteen selected metabolites underwent metabolic pathway analysis and were evaluated for their potential as biomarkers. Three of these metabolites showed potential as biomarkers (AUC>0.65): oxaloacetate and 5′-phosphoribosyl-N-formylglycinamidine, which decreased in the uncontrolled HbA1c group, and (S)-dihydroorotate, which increased in the uncontrolled HbA1c group. The involved metabolic pathways included (1) alanine, aspartate, and glutamate, (2) citric acid (Krebs cycle), (3) gluconeogenesis, (4) pyruvate, (5) pyrimidine, and (6) purine.
Based on these results, there’s a significant difference in urine metabolites of the two groups that potentially be the biomarkers for DKD progression with controlled and uncontrolled HbA1c.
Type 2 diabetes mellitus, Diabetic kidney disease, Metabolomic study, Biomarker identification, Pathway analysis.
Diabetes mellitus (DM) is a chronic metabolic disorder characterized by persistent hyperglycemia. This condition arises due to impaired insulin secretion, insulin resistance in peripheral tissues, or combination of both conditions.1 According to the International Diabetes Federation (IDF), approximately 19.5 million adults in Indonesia are affected by diabetes, ranking the country fifth highest globally in diabetes prevalence.2 Notably, 90-95% of these cases are Type 2 Diabetes Mellitus (T2DM), which is largely preventable through lifestyle modifications.3
Persistent hyperglycemia in individuals with diabetes mellitus (DM) can lead to multi-organ damage and subsequent complications thus underscoring the necessity for effective preventive strategies. One significant complication that affects 30-40% of diabetic patients is renal dysfunction.4 This condition is termed Diabetic Kidney Disease (DKD) and is associated with a high risk of mortality. It may progress to End-Stage Renal Disease (ESRD), the terminal phase of kidney function impairment.4,5 Intensive glycemic control can prevent both the onset and development of diabetes complications.1 The standard therapeutic approach for managing T2DM often involves the use of Oral Antidiabetic Drugs (OAD) such as metformin.6 T2DM patients who have been treated with single OAD but still have not reached the HbA1c target (≤7%) within three months, can be given combination OAD.6 Metformin is frequently administered in combination with glimepiride to enhance its efficacy. The rationale for combining metformin with glimepiride stems from their complementary mechanisms of action, metformin enhances insulin sensitivity and decreases hepatic glucose production, while glimepiride promotes pancreatic insulin secretion.7
Early DKD is asymptomatic and often difficult to detect.8 The Estimated Glomerular Filtration Rate (eGFR) and Urine Albumin-Creatinine Ratio (UACR) are commonly used biomarkers for detecting and staging kidney function impairment.5 However, these biomarkers have limitations. Kidney damage may precede the detection of albuminuria, as observed in normoalbuminuric patients.9 Furthermore, eGFR can decline due to various factors such as age, sex, pregnancy, unusual muscle mass, cirrhosis, nephrotic syndrome, a past solid organ transplant, and some medications.8 Beyond renal function, it is crucial to consider HbA1c levels in DKD patients. Uncontrolled HbA1c (>7%) can influence the progression of DKD as there is a significant negative correlation between HbA1c levels and estimated glomerular filtration rate (eGFR). HbA1c variability is significantly associated with an increased risk of albuminuria.10 Consequently, there is a need for alternative biomarkers that offer greater accuracy in diagnosing DKD, evaluating therapeutic efficacy, and monitoring disease progression, thereby facilitating earlier detection and timely intervention to improve patient outcomes.
Metabolomics study is currently a prominent method used to identify metabolites by analyzing the end products of biochemical processes in the body. Changes in metabolite composition contribute to unravelling the pathophysiological and physiological conditions of chronic diseases, as it operates at the molecular level, which facilitates the discovery of disease biomarkers.11,12 Liquid Chromatography Quadrupole Time-of-Flight Mass Spectrometry (LC/MS-QTOF) is the most widely employed analytical strategy for metabolomics, especially untargeted metabolomics, considering its wide metabolite coverage and high sensitivity.13 The identified compounds can reveal novel metabolic pathways from the changes in metabolic profiles that can be the new potential biomarkers for diagnosing and evaluating DKD.14
Therefore, it is essential to perform an untargeted metabolomics study by differentiating between those with controlled and uncontrolled HbA1c levels in conducting a comparative analysis of the urine profiles of T2DM patients with low-risk DKD. This study focuses on patients with a low risk of DKD to enhance early detection before the disease progresses to more severe stages.
Ethical approval for this study was obtained from the FKUI-RSCM Health Research Ethics Committee, Universitas Indonesia (No. KET-364/UN2.F1/ETIK/PPM.00.02/2023 and No. KET-245/UN2.F1/ETIK/PPM.00.02/2024) and the South Jakarta Health Administration Sub-Department. The study was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained and signed by all participants prior to their inclusion in the study.
The research was conducted as an observational study using a cross-sectional design. A total of 32 samples were obtained from Pasar Minggu District Health Center in February 2023–April 2024. The samples were divided into two groups based on their HbA1c levels: controlled HbA1c (≤7%) (n=16) and uncontrolled HbA1c (>7%) (n=16). Blood samples were collected for HbA1c and eGFR measurements, while urine samples were analyzed for UACR and metabolites. Subject participation was obtained through the signing of informed consent after a thorough explanation of the study. Sample collection was carried out using the consecutive sampling technique, wherein subjects meeting the predefined inclusion criteria were selected and included until the required sample size was achieved. The inclusion criteria included (1) age ≥36 years old, (2) treatment of metformin-glimepiride for at least six months, and (3) included in the low-risk category for diabetes kidney disease based on KDIGO 2022. Exclusion criteria included (1) no urine and blood specimens were obtained; (2) pregnant and lactating women; (3) severe anemia with or without receiving blood transfusions; (4) stroke, chronic liver disease, congestive heart failure, and infectious diseases.
The HbA1c and UACR levels were measured using Afinion™ AS100 Analyzer (Alere Technologies; Serial number: SNAS0044016, USA). The analysis results will be visible on the tool screen, showing the HbA1c levels in (%) unit and UACR levels in (mg/g) unit.
Midstream urine samples were collected and subsequently centrifuged at 5,000 × g for 30 minutes at 4°C (ThermoFisher Scientific, USA). The supernatant was filtered through a 0.2-micron syringe filter (Membrane Solutions, USA) and the filtrate was added with 100 μL of 100 mM sodium azide (Merck, Germany) as the preservative agent. After that, samples were immediately stored at -80°C (Sanyo, Japan).
A volume of 100 μL of the thawed sample was transferred to a 1.5 mL microtube (OneMed, Indonesia) using a micropipette (ThermoFisher Scientific, USA). Subsequently, 400 μL of 100% methanol (Merck, Germany) was added to the urine sample in the microtube. The solution was vortexed for 30 seconds (Bionex, USA) and then incubated at -20°C for 15 minutes (Nihon, Japan). The samples were centrifuged at 10,000 rpm for 15 minutes at 4°C (Eppendorf, Germany). The supernatant was transferred to a new 1.5 mL microtube (OneMed, Indonesia) and then dried using a vacuum concentrator (Tomy, USA) to obtain a dry powder of urine sample. The dried powder was stored at -80°C (Sanyo, Japan) until metabolomic analysis using LC/MS-QTOF (6520 Agilent Technologies, SA, USA). Catalogue numbers were recorded where available; when not applicable, reagents were identified by supplier and purity grade.
The urine samples were reconstituted with 30 μL of mobile phase solvent A (0.1% formic acid in dH2O) and solvent B (0.1% formic acid in ACN), vortexed, and centrifuged at 10,000 rpm for 15 minutes at 4°C (Eppendorf, Germany). The approximately 20 μL of supernatant was collected and ready for injection and analysis. Injection of 2 μL of the sample was carried out with LC/MS-QTOF (model 6520 Agilent Technologies, SA, USA) using a ZORBAX Eclipse Plus C18 column (Agilent Technologies, SA, USA) maintained at 40°C. The flow rate was 0.25 mL/minute with a linear gradient comprising solvent A and solvent B over 18 minutes from 5% to 95% of Mobile Phase B. Then, for 12 minutes 95% of Mobile Phase B was maintained for flushing after each injection. The electrospray ionization (ESI) source with standard settings were as follows: V Cap 4000 V, skimmer voltage 68 V, and fragment 215 V. The nebulizer was set at 30 psi, and the nitrogen drying gas was set at a flow rate of 10 L/min at a temperature of 325°C. Data were collected via positive ESI mode with full scan mode from 50 to 1000 mass/charge (m/z). The reference masses used are 121.0509 m/z (C5H4N4) and 922.0098 m/z (C18H18O6N3P3F24) to allow mass corrections accurately and consistently.
Each of the urine samples was run separately. Small aliquots of the whole samples were pooled for Quality Control (QC) to ensure the accuracy and reproducibility of the analysis method. In each batch, eight samples were run with one blank and one QC. The QC was injected at the beginning, middle, and end of the analysis to confirm the system stability and performance. Then, QC samples were evaluated by calculating the Relative Standard Deviation (RSD) of selected metabolites that appeared consistently in each batch of QC sample.15
Raw data and metabolite extraction were processed using Agilent Technologies MassHunter Qualitative software. For the positive ionization mode, the adducts used are H+ and Na+. Metabolites were detected within 0.100 to 18,000 min and 50 to 1000 m/z of the analysis. Then, the raw data was converted using the MSConvert ProteoWizard software.
Statistical analysis, metabolic pathway mapping, and determination of potential biomarkers were performed using the open-source web-based software MetaboAnalyst 6.0. The identified metabolites were annotated using the Human Metabolome Database (HMDB), METLIN, PubChem, and Kyoto Encyclopedia of Genes and Genomes (KEGG). The potential biomarkers were determined using the Area Under Receiver Operating Characteristics Curve (AUROC) analysis.
IBM SPSS Statistics was used for statistical analysis. Normal distributed numerical (quantitative) data are expressed as mean ± standard deviation (SD), and non-normal distributed data are expressed as median (minimum-maximum). Meanwhile, categorical (qualitative) data are expressed as proportions or frequency of events. Differences between groups were compared using the independent t-test for normal distributed data and the Mann-Whitney test for non-normal distributed data. Comparisons of categorical data were compared using the Chi-square test.
A total of 1,772 Type 2 Diabetes Mellitus (T2DM) patients from Puskemas Pasar Minggu were assessed during the February-April recruitment period. After screening, 37 patients met the inclusion and exclusion criteria and were enrolled as participants. The primary reasons for exclusion from the initial pool were: having an infection (n=1), subject refusal (n=2), falling into a moderate DKD risk category (n=9), and falling into a high DKD risk category (n=2).
From the 37 enrolled participants, 32 successfully provided both blood and urine samples. Five participants were excluded at this stage: three were absent, and urine samples could not be obtained from two. These 32 participants constituted our final study sample.
During laboratory analysis, 2 urine samples could not be analyzed, resulting in 30 samples being processed with LC/MS-QTOF at iPROMISE. The final distribution for metabolomic analysis was 16 samples in the low-risk group, comprising 9 with controlled HbA1c (≤7%) and 7 with uncontrolled HbA1c (>7%). A flow diagram of participant selection is presented in Figure 1.
Complete clinical and demographic data were available for all 32 participants included in the final analysis. No missing data were encountered for any variables of interest presented in Table 1 and Table 2. The basic characteristics observed in the study were age, sex, body mass index (BMI), patient compliance, exercise and physical activity, smoking habits, metformin therapy regimen, glimepiride therapy regimen, comorbid conditions, and the duration of T2DM of each subject as shown in Table 1. The clinical characteristics observed in the study were blood pressure, UACR, and eGFR as shown in Table 2.
| Characteristics | HbA1c Levels | p | |
|---|---|---|---|
| Controlled (n=16) | Uncontrolled (n=16) | ||
| Gender | |||
| Male | 7 (43.8) | 4 (25) | 0.457a |
| Female | 9 (56.3) | 12 (75) | |
| Age | |||
| <65 years | 11 (68.8) | 10 (62.5) | 1.000a |
| ≥65 years | 5 (31.3) | 6 (37.5) | |
| Body Mass Index (BMI) | |||
| BMI <18.5 | 0 (0) | 1 (6.3) | 0.229b |
| BMI 18.5–22.9 | 2 (12.5) | 5 (31.3) | |
| BMI ≥23.0 | 14 (87.5) | 10 (62.5) | |
| History of T2DM | |||
| <5 years | 4 (25) | 2 (12.5) | 0.564b |
| ≥5 years | 12 (75) | 14 (87.5) | |
| Exercise routine | |||
| Yes | 5 (31.3) | 5 (31.3) | 1.000a |
| No | 11 (68.8) | 11 (68.8) | |
| Smoking habits | |||
| Yes | 13 (81.3) | 15 (93.8) | 0.600b |
| No | 3 (18.8) | 1 (6.3) | |
| Other diseases | |||
| Yes | 3 (18.8) | 2 (12.5) | 1.000a |
| No | 13 (81.3) | 14 (87.5) | |
| Metformin therapy regimen | |||
| 1 × 500 mg | 3 (18.8) | 4 (25) | 0.904a |
| 2 × 500 mg | 4 (25) | 4 (25) | |
| 3 × 500 mg | 9 (56.3) | 8 (50) | |
| Glimepiride therapy regimen | |||
| 1 × 2 mg | 12 (75) | 15 (93.8) | 0.264a |
| 1 × 4 mg | 2 (12.5) | 1 (6.3) | |
| 2 × 2 mg | 2 (12.5) | 0 (0) | |
| Medication adherence | |||
| Yes | 8 (50) | 4 (25) | 0.273b |
| No | 8 (50) | 12 (75) | |
| Characteristics | HbA1c Levels | p | |
|---|---|---|---|
| Controlled (n=16) | Uncontrolled (n=16) | ||
| Systole blood pressure (mmHg) | 130 (110–148) | 130 (108–197) | 0.662b |
| Diastole blood pressure (mmHg) | 70 (66–89) | 70 (51–84) | 0.495b |
| Urine Albumin to Creatinine Ratio (UACR) | 1.65 ± 0.15 | 1.40 ± 0.13 | 0.344a |
| Estimated Glomerulus Filtration Rate (eGFR) | 87.06 ± 3.40 | 88.75 ± 3.50 | 0.651a |
Both characteristics show no significant differences between the controlled HbA1c and uncontrolled HbA1c groups (p>0.05). This result indicates that the confounding variables were equivalent, allowing for a more accurate comparison of both groups.
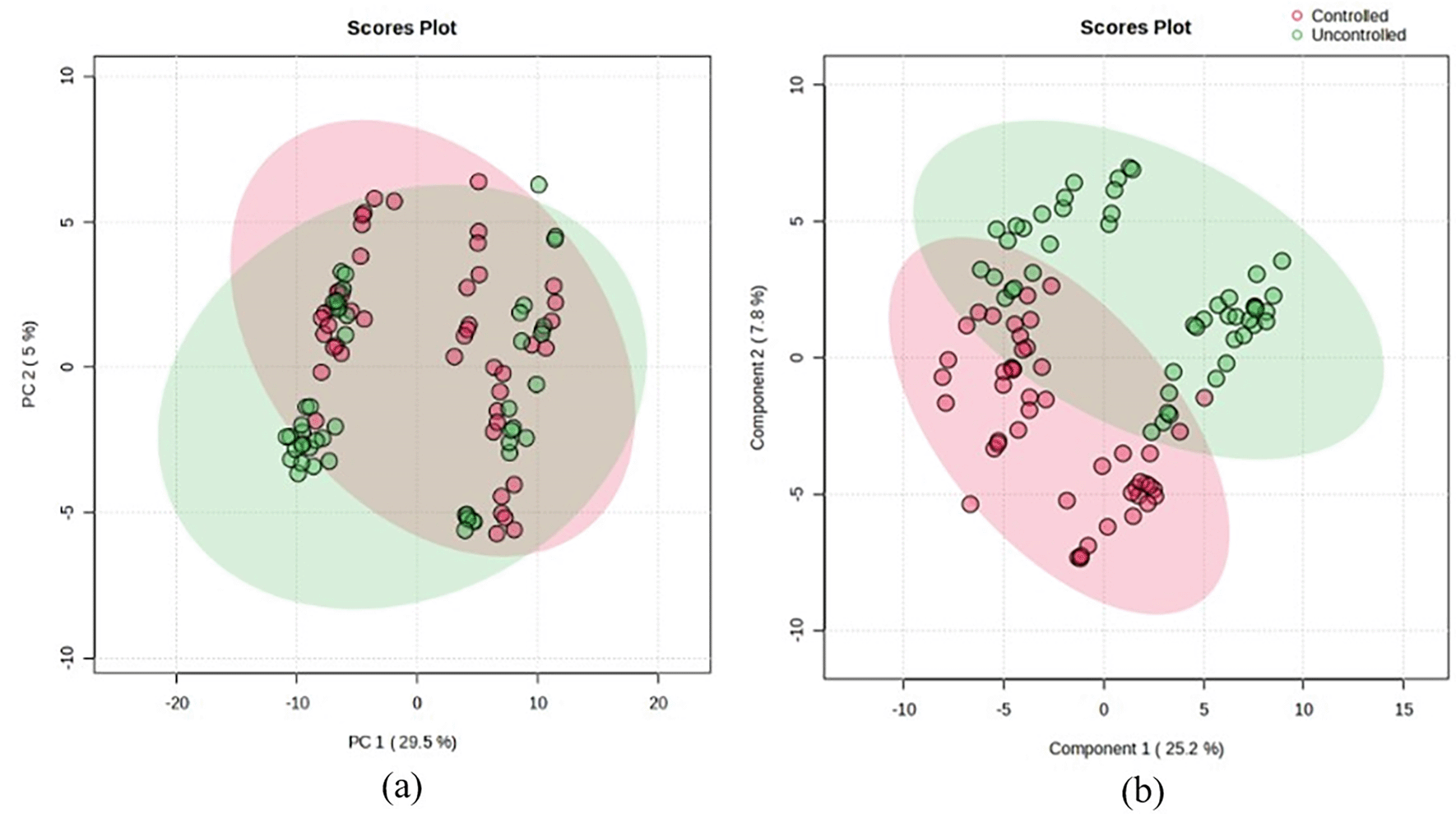
Multivariate analysis was conducted using Principal Component Analysis (PCA) and Partial Least Squares-Discriminant Analysis (PLS-DA) as the data clustering visualization of the two groups, as shown in Figure 2. PCA and PLS-DA score plots are commonly used multivariate statistical methods in metabolomics analysis. PCA groups samples with similar metabolite profiles into a pattern or cluster.16 While PCA provides a general representation of the data structure, PLS-DA focuses on the clustering or separation of the analyzed sample groups. The plots indicate that the two groups are well-separated, with all samples clustering according to their respective groups, particularly in the PLS-DA analysis. Unlike PLS-DA, which is a supervised approach, PCA is unsupervised, resulting in more distinct clustering in PLS-DA score plots compared to PCA.
In addition to PCA and PLS-DA, a heatmap is also utilized as a visualization method in metabolomic analysis. This method uses colour intensity to indicate the abundance of detected metabolites in each sample group. The heatmap shown in Figure 3 provides a detailed overview of metabolite abundance profiles within each experimental group.
The significance of metabolite differences was evaluated and selected based on the criteria of VIP>1, p-value<0.05, and log2(FC)>1.2 which were shown in Figure 5.17,18 The VIP score was obtained from PLS-DA analysis and served as a parameter describing the contribution of metabolites to each group. Metabolites with a VIP score greater than 1 were considered to have made a significant contribution ( Figure 4). Meanwhile, log2(FC) represented the log transformation of the fold change in metabolite abundance between study groups. A log2(FC) value greater than 1.2 indicated that the average metabolite abundance was 1.2 times higher or lower in one group compared to another.17 Metabolite abundance is either upregulated or downregulated in the sample group, according to positive and negative results on the log 2-fold change value (Log2(FC)).
Following the initial screening based on VIP score, p-value, and log2(FC) criteria, differential metabolites were identified using the HMDB, METLIN, Metabolomic Workbench, and PubChem databases. The significant metabolites identified by VIP score analysis were then used for biochemical pathway analysis. The diagnostic performance of the selected classifiers was evaluated using ROC implemented in MetaboAnalyst 6.0.
From the previous metabolites screening, 15 metabolites met the criteria and were further analyzed to identify the biochemical pathways involved for each metabolite and their potential as biomarkers. The significance of the pathways was illustrated by the position of circles in the pathway analysis graph, where those farther to the right indicated a higher impact ( Figure 6). Each circle in the graph represented a pathway involved in the metabolism of influential metabolites, and from the graph, it was found that the most significant metabolic pathway was the citric acid cycle (Krebs cycle).
Based on the pathway analysis results, the involved metabolites were identified: oxaloacetate, which is involved in the (1) alanine, aspartate, and glutamate metabolism, (2) citric acid cycle (Krebs cycle), (3) gluconeogenesis, and (4) pyruvate metabolism; acetylphosphate, which is involved in pyruvate metabolism; 3-phosphonooxypyruvate, which is involved in the (1) cysteine and methionine metabolism, and (2) glycine, serine, and threonine metabolism; (5′-phosphoribosyl)-N-formylglycinamidine, which is involved in purine metabolism; and (S)-dihydroorotate, which is involved in pyrimidine metabolism. These five metabolites were significantly different between the two groups and thus have the potential to serve as biomarkers.
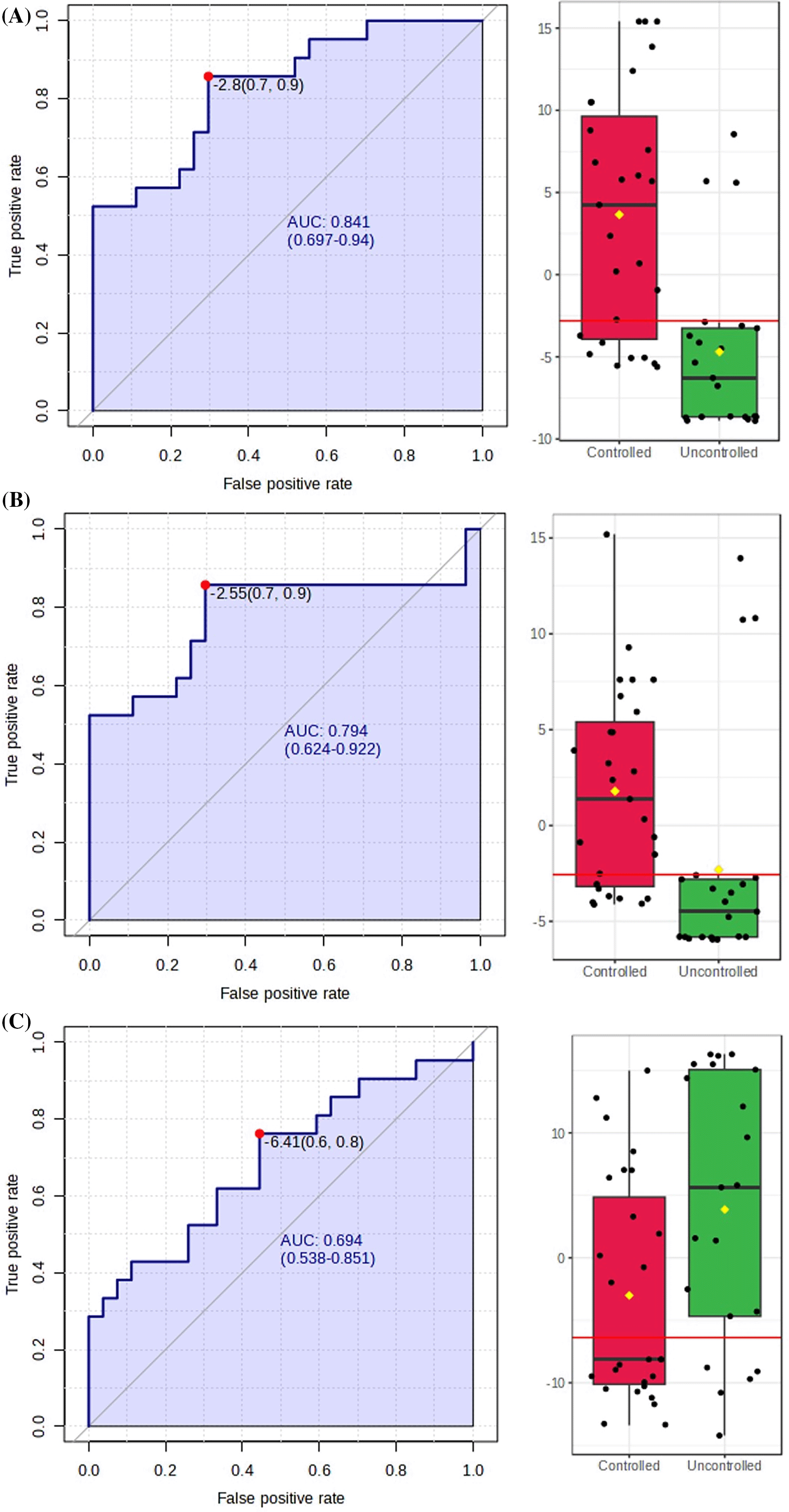
Further analysis with the Area Under the Receiver Operating Characteristic (AUROC) curve was needed to assess their potential as biomarkers as shown in Figure 7. Metabolites with biomarker potential were visualized by an increasing true positive rate and a decreasing false positive rate.15 The potential biomarkers must meet diagnostic efficiency criteria with an AUC>0.65.18 The larger AUC value, the higher the predictive accuracy, meaning that the closer the curve is to the upper left corner, the higher the predictive accuracy.19 The maximum AUC value is 1, indicating no overlap in test data between the two groups, making the metabolite a highly significant potential biomarker.
The previous five metabolites identified through pathway analysis were narrowed down to three based on their AUC values. The final three metabolites that met the criteria and showed significant potential as biomarkers are oxaloacetate, (5′-phosphoribosyl)-N-formylglycinamidine, and (S)-dihydroorotate listed in Table 3. A boxplot visualization, shown in Figure 7, demonstrates that the metabolites do not overlap between the groups. The boxplot results are also reflected in the AUC values for each potential biomarker, with a higher AUC corresponding to better boxplot visualization.
Metabolic impairment in diabetes mellitus affects the synthesis and degradation of oxaloacetate. The hyperglycaemic state experienced by diabetic patients led to an increased influx of glucose into the non-insulin-dependent cells, which enhanced the activity of the gluconeogenesis pathway and the Krebs cycle. This process resulted in an overproduction of NADH and FADH2, which entered the mitochondrial electron transport chain.20 Such excessive production triggered the formation of free radicals, leading to oxidative stress that caused cellular damage, including in renal cells, ultimately contributing to the development of DKD. In this condition, an upregulation of gluconeogenesis and disruption of the Krebs cycle occurred, potentially causing oxaloacetate degradation due to its increased utilization in gluconeogenesis and impaired regeneration through the Krebs cycle.20 The metabolite (5′-phosphoribosyl)-N-formylglycinamidine (FGAM) was involved in de novo purine metabolism. Increased free radical production and oxidative stress in DKD conditions could damage enzymes involved in the purine biosynthesis pathway, including FGAM synthetase. This damage disrupted purine production, which plays a crucial role in DNA and RNA synthesis, further exacerbating cellular damage in the kidneys. A reduction in this metabolite could serve as a marker for the presence of DKD. The (S)-dihydroorotate metabolite plays a role in the pyrimidine biosynthesis pathway. Mitochondrial dysfunction was identified as a potential risk factor for kidney disorders since the kidneys rely on mitochondria to provide energy for various cellular functions such as excretion, nutrient reabsorption, fluid balance, and blood pressure regulation.21 Under hyperglycemic conditions, renal cells experience metabolic stress that disrupts mitochondrial function and associated metabolic pathways.22 In this setting, pyrimidine metabolism, including dihydroorotate-related pathways, likely plays an important role in cellular adaptation. Therefore, altered dihydroorotate levels observed in this study may reflect an adaptive metabolic response of renal cells to hyperglycemia-induced mitochondrial stress, supporting its potential relevance as an early metabolic biomarker in DKD.
Future studies involving a larger patient sample size could enhance the robustness of the metabolite findings. Improved metabolite identification may be achieved by utilizing more advanced software, online tools, or comprehensive metabolite databases, allowing for a more holistic analysis and minimizing potential bias. Furthermore, this study has several limitations. The cross-sectional design prevents inference of causality and the relatively small sample size may lead to imprecision in the estimates. The use of consecutive sampling from a single primary health center may limit the representativeness of our sample and introduce selection bias. Consequently, the generalizability (external validity) of our findings is likely limited to T2DM populations with similar characteristics, such as adults with low-risk DKD who are undergoing combination therapy with metformin and glimepiride in a similar primary care setting in Indonesia. External validation in broader and more diverse populations is necessary to confirm the wider applicability of these potential biomarkers.
There are three metabolites with significant potential to serve as biomarkers for low-risk diabetic kidney disease: oxaloacetate, 5′-phosphoribosyl-N-formylglycinamidine, which decreased in the uncontrolled HbA1c group, and (S)-dihydroorotate, which increased in the controlled HbA1c group. The metabolic pathways involved include (1) alanine, aspartate, and glutamate, (2) citric acid (Krebs cycle), (3) gluconeogenesis, (4) pyruvate, (5) pyrimidine, and (6) purine. To elucidate a complete understanding of the significance of these potential biomarkers in the progression of diabetic kidney disease (DKD), further research employing a targeted metabolomic approach is required.
Zenodo: Urine Metabolomic Analysis of Type 2 Diabetes Mellitus Patients with Controlled and Uncontrolled HbA1c in Low-Risk Diabetic Kidney Disease. https://doi.org/10.5281/zenodo.17556311.23
The project contains the following underlying data:
• T2DM_Urine_Metabolomics_Dataset.xlsx (Anonymized clinical data and the annotated peaklist) Clinical_data.xlsx (anonymized clinical data including HbA1c, eGFR, and UACR values) and Annotated_peaklist.xlsx (LC/MS-QTOF metabolite peaklist processed through MetaboAnalyst 6.0)
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Zenodo: Urine Metabolomic Analysis of Type 2 Diabetes Mellitus Patients with Controlled and Uncontrolled HbA1c in Low-Risk Diabetic Kidney Disease. https://doi.org/10.5281/zenodo.17556311.23
This project contains the following extended data:
• Informed_Consent_Template_English.pdf (Informed consent form used in recruitment)
• Questionnaire_Form_English.pdf (Questionnaire form used in recruitment)
• Supplementary_Figure_1.tiff (Participants Recruitment Flowchart)
• Supplementary_Figure_2.tiff (PCA and PLS-DA score plots)
• Supplementary_Figure_3.tiff (Heatmap visualization)
• Supplementary_Figure_4.tiff (VIP Scores plot)
• Supplementary_Figure_5.tiff (Urine Metabolites Screening between Controlled and Uncontrolled HbA1c Groups)
• Supplementary_Figure_6.tiff (Pathway impact analysis)
• Supplementary_Figure_7A-C.tiff (AUROC and boxplot analysis)
• Supplementary_Table_1.xlsx (List of significant metabolites with p-value, VIP, log2FC and associated pathways)
• Supplementary_Table_2.xlsx (Basic characteristics of study participants)
• Supplementary_Table_3.xlsx (Clinical characteristics of study participants)
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Zenodo: STROBE checklist for “Urine Metabolomic Analysis of Type 2 Diabetes Mellitus Patients with Controlled and Uncontrolled HbA1c in Low-Risk Diabetic Kidney Disease.” https://doi.org/10.5281/zenodo.17471280.24
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)