Introduction
Transcription factors regulate gene expression in the mammalian brain, playing a critical role in both neurodevelopment and in neuronal plasticity during its lifespan. During development, transcription factor mediated regulation is essential for appropriate cell fate specification, cell migration and connectivity1–3. Transcription factors also regulate plasticity including activity-dependent process of dendritic pruning, axonal sprouting and cell proliferation and survival4–6.
One family of transcription factors, the LIM-homeodomain (LIM-HD) family, is known to play critical roles in regulating cell proliferation, axon outgrowth and pathfinding across several systems7–11. The LIM-HD proteins have a C-terminal homeodomain which binds to DNA and two zinc finger “LIM” domains that bind co-factors encoded by the Clim genes. The transcriptionally active complex is a tetramer comprising two LIM-HD molecules bridged by a dimer of two Clim molecules12,13. LIM-only (Lmo) proteins lack the homeodomain but can bind Clim molecules, and function as dominant-negative regulators of LIM-HD function12,14,15. At least thirteen LIM-HD (Lhx) genes four Lmo genes and two Clim genes have been identified in the mouse. A subset of genes is expressed in the embryonic and mature hippocampus and of these, Lhx2 and Lhx5 are critical to hippocampal development16. Lhx2 plays a fundamental role in early telencephalic development as a cortical selector gene11. The neocortex and hippocampus do not form in the absence of Lhx211,17. At later stages, Lhx2 plays a new role in the developing hippocampus, as a necessary and sufficient repressor of astrogliogenesis18. Lhx2 continues to be expressed in the mature hippocampus. Lhx5 is critical for hippocampal development at early stages, but is not expressed in the embryonic hippocampus once it is specified19. Lhx1, Lhx9, Clim1a, Clim2, Lmo3 and Lmo4 are all expressed in the hippocampus at embryonic and adult stages16, but no loss of function phenotypes have been reported in the hippocampus.
While several studies have implicated the LIM-HD family as a key modulator of important neurodevelopmental events, the understanding of the role of this transcription factor family in the postnatal and adult brain remains relatively unexplored. These transcription factors are known to regulate cell proliferation8,20, axon pathfinding21,22 and neurite outgrowth23,24. These phenomena have parallels in the structural plasticity that occurs in postnatal and adult life. It is now well established that the same molecules that bring about the early development of the hippocampus are often reutilized in adult reorganization and structural plasticity25–27. Several LIM-HD family members continue to be expressed in the adult hippocampus16 (this study). Therefore, we explored whether these genes display activity-dependent regulation in the adult hippocampus, to provide a basis for studies that may uncover new functions for these genes in maturity.
Activity dependent neuronal plasticity has been suggested to reutilize key developmental pathways to evoke plasticity in the mature nervous system. In particular, seizure models have been shown to induce dramatic changes in progenitor proliferation, axonal sprouting, dendritic reorganization, changes in neuronal cell survival and progenitor differentiation within the hippocampus28–31. Intriguingly, the nature of neuroplastic changes evoked by seizures differs quite dramatically in the postnatal versus the adult brain32–34. Regulation at the level of signaling and transcription factors has been shown to be important for structural plasticity in the hippocampus35. While neuronal activity and seizures are likely to recruit major developmental signalling pathways in the hippocampus, thus far the role of key developmental transcription factor families as targets is relatively unexplored.
An earlier study reported that LIM-only genes Lmo1, 2 and 3 are differentially regulated in a field-specific manner in the adult rat hippocampus in response to kainic acid-induced seizure36. We examined a broader set of Lmo and LIM-HD genes as well as their co-factors in a similar paradigm, not only in the adult rat hippocampus, but also in early postnatal stages when hippocampal circuitry is not fully developed37–39. Our study provides evidence that LIM-HD, LIM-only, and Clim gene mRNA displays selective field-specific regulation in the hippocampus in response to kainate induced seizures. This provides a basis to explore potential new functions of these genes in activity-dependent synaptic plasticity.
Results
In this study we focused on LIM-HD genes that are expressed in the adult hippocampus, Lhx1, Lhx2 and Lhx9 and their co-factors, Clim1a and Clim2. Among the LIM-only genes, Lmo1, Lmo2, and Lmo3 have been previously reported to display differential regulation in kainate-induced seizure36. In our study, we included Lmo3 as a control to allow comparison with the earlier study36, and also Lmo4 which was not examined previously. We examined the mRNA expression of these genes at postnatal day P7 when the hippocampal circuitry is not yet fully developed, and also in adult rats (2–3 months old) with mature hippocampal neurocircuitry.
Differential expression of LIM family members and their co-factors across different hippocampal fields
We used non-radioactive in-situ hybridization to examine gene expression in the CA1 and CA3 fields of the Ammon’s horn as well as the dentate gyrus (DG) of control animals (Figure 1a). Lhx1 transcripts were not detectable in the hippocampus at P7, and only weakly expressed in the adult DG (Figure 1b). In contrast, Lhx2 and Lhx9 are expressed intensely in the DG and CA3, with weaker expression in CA1 at P7. In the adult, expression was strong in the DG, but weak in CA1 and CA3 (Figure 1c, d). Lmo3 and Clim2 are strongly expressed in CA1 and DG, with weaker expression in the CA3 region at both stages (Figure 1e, h). Lmo4 shows strong expression in CA1 but is weakly expressed in CA3 and DG at both stages (Figure 1f). Clim1a displays expression in all fields at P7, but is weak to undetectable in CA3 in the adult (Figure 1g).
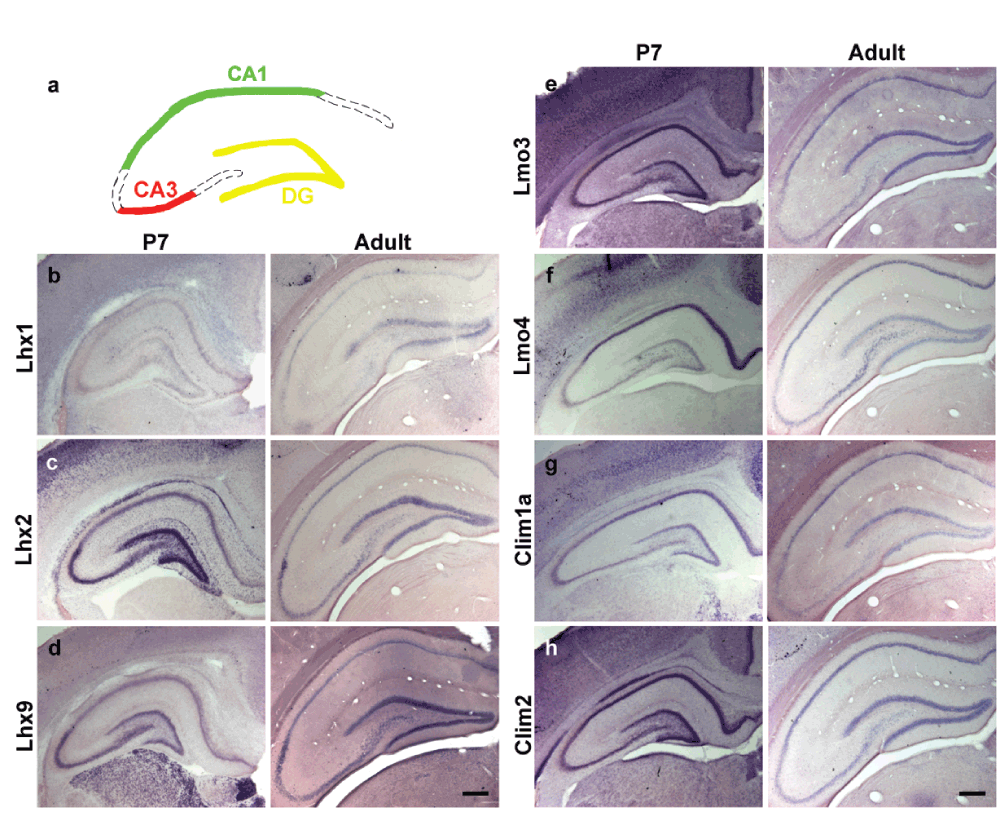
Figure 1. Expression of LIM genes and co-factors in early postnatal and adult hippocampus.
(a) A schematic illustrating hippocampal subfields; dentate gyrus (DG) (yellow), CA3 (red) and CA1 (green) fields of Ammon’s horns. (b–d) Non-radioactive in-situ hybridization of LIM-homeodomain genes at postnatal day (P)7 and in adult control animals showing differential expression of LIM-homeodomain genes; Lhx1 (b), Lhx2 (c) and Lhx9 (d), Lmo3 (e), Lmo4 (f) Clim1a (g), Clim2 (h) across the hippocampal subfields. Scale bars = 200µm
Activity is known to regulate structural plasticity and neurogenesis in the adult hippocampus40,41. We administered kainate intraperitoneally to both early postnatal and adult rats to induce seizures as a model of activity and analysed whether there is differential regulation of LIM genes in response to kainate-evoked seizures 6 hours later. All animals administered kainate exhibited classical hallmarks of seizure. Using radioactive in-situ hybridization and optical densitometry we assessed the expression of Lhx1, Lhx2, Lhx9, Lmo3, Lmo4, Clim1a and Clim2 in the postnatal and adult hippocampal subfields (see Materials and methods). Radioactive in-situ hybridization has an important advantage over quantitative PCR since it provides spatial resolution. The hippocampal CA1 and CA3 fields are molecularly distinct, and the dentate gyrus contains a distinct cell population from the Ammon’s horn42. Therefore it is necessary to quantitate the gene expression in each region individually.
Seizure induced regulation of LIM family members and their co-factors in the adult dentate gyrus (DG)
The DG displays robust structural changes in response to seizure. Increase in dentate granule cell neurogenesis43 and extensive mossy fiber sprouting41,44 are hallmarks of kainate induced seizure. Upon kainate treatment, the expression of Lhx1 showed a striking increase (25%; p = 0.019) in the adult DG. This is in contrast with the adult Lhx2 and Lhx9 expression, the mRNA levels of which show a drastic reduction (60% for Lhx2, p = 0.0004 and 36% for Lhx9, p = 0.003; Figure 2, Figure 3a). Interestingly, the LIM-only genes Lmo3 and Lmo4 also showed opposite changes: whereas Lmo3 levels decreased significantly (53%, p = 0.002), Lmo4 mRNA levels showed a remarkable increase (55%, p = 0.009) in kainate-treated animals. The decrease in Lmo3 levels was consistent with that reported previously36. The mRNA levels of the cofactor Clim1a decreased slightly in treated animals (15%, p = 0.048) whereas no significant difference was observed with Clim2 (Figure 2, Figure 3b).
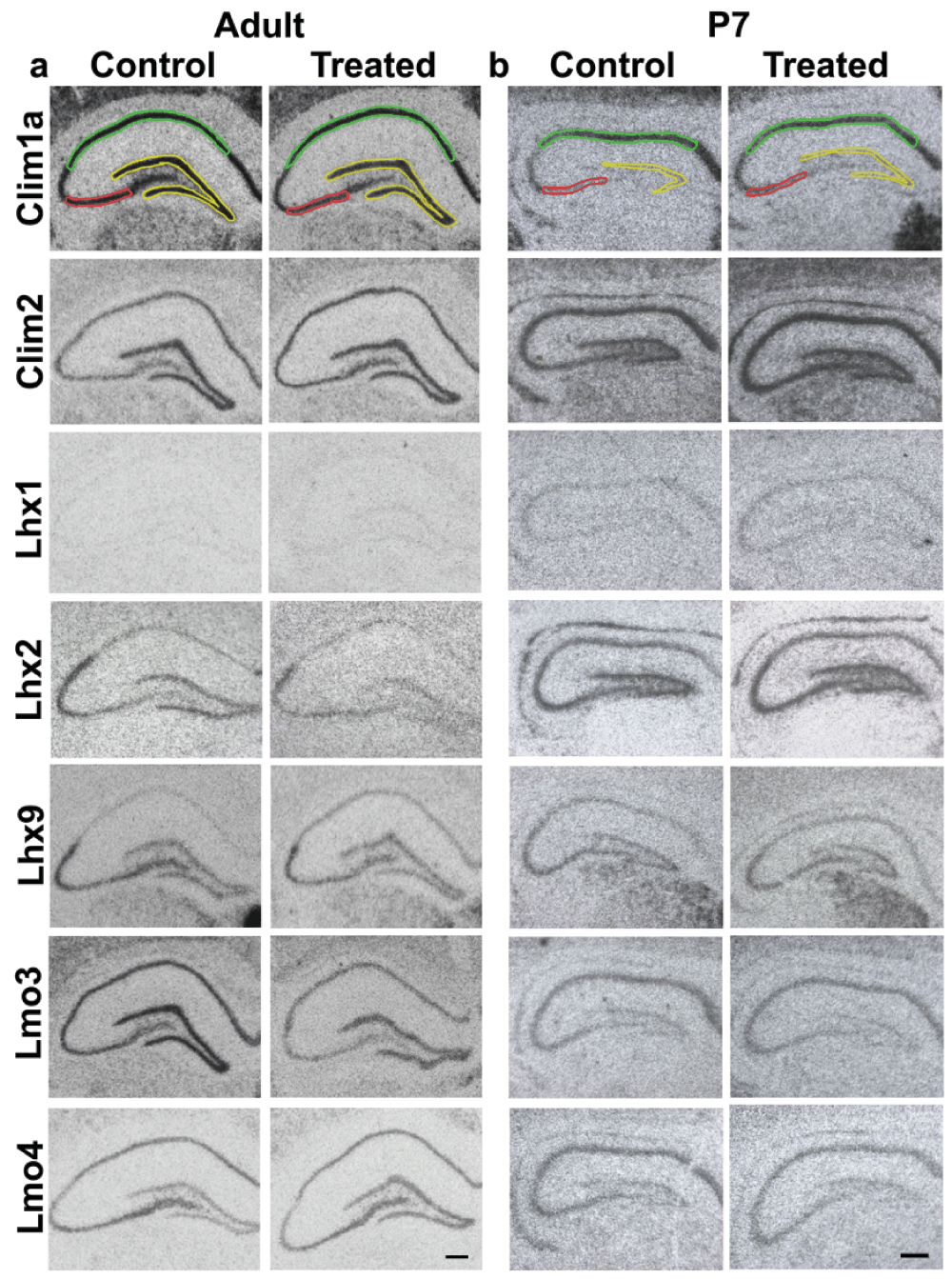
Figure 2. Expression of LIM genes and co-factors used for densitometric analysis.
Representative images of sections of brains from control and kainate-administered animals processed for radioactive in-situ hybridization of LIM-homeodomain genes in the hippocampus. 1 section from each condition is shown for adult (a) and P7 (b) animals. Colored lines mark the areas for quantification of expression in different hippocampal subfields: DG (yellow); CA3 (red); CA1 (green). Scale bars = 200µm.
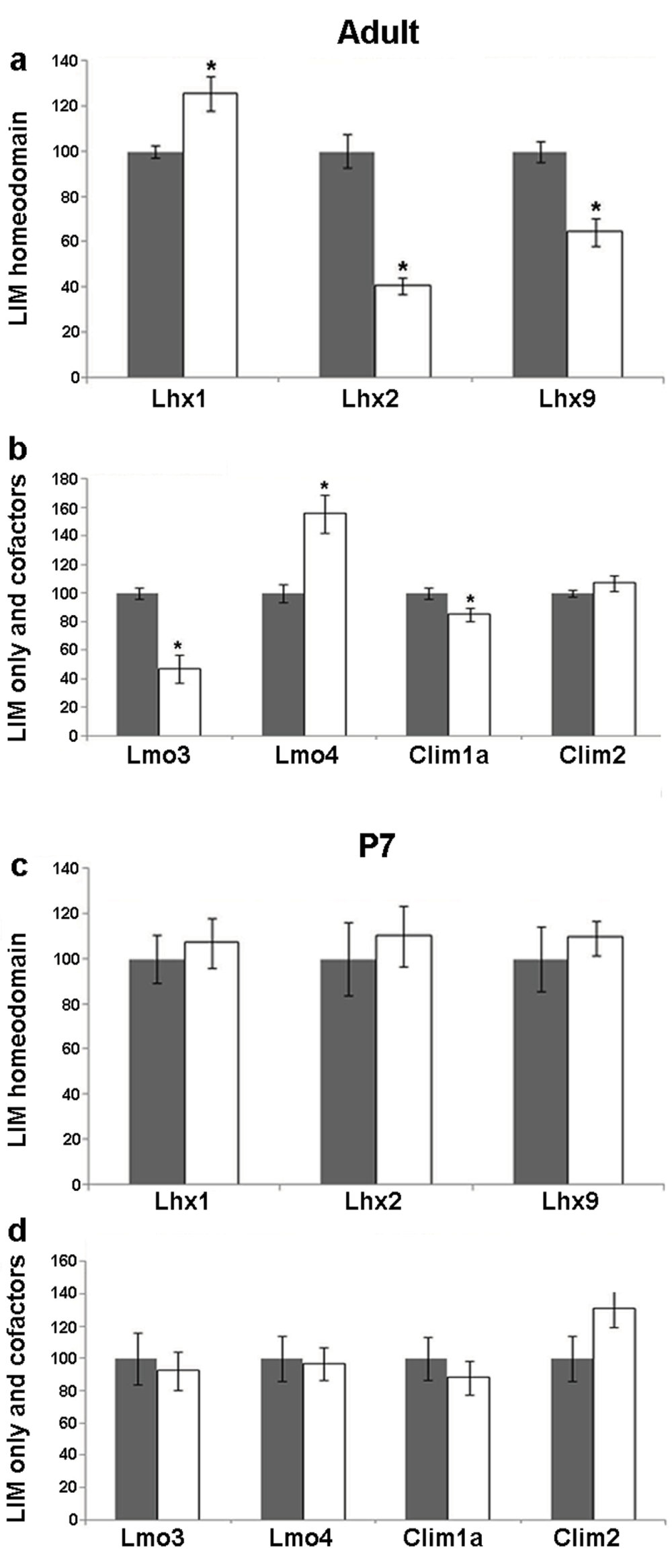
Figure 3. Kainate-induced regulation of LIM genes and co-factors in the dentate gyrus (DG) of adult and P7 rats.
Quantitative densitometric analysis of the adult DG region following kainate administration in adult (a,b) and P7 (c,d) rats. Grey bars are controls, white bars are kainate treated animals. Results are expressed as mean ± SEM percentage of control for mRNA expression (*p < 0.05, unpaired Student’s t test).
Seizure induced regulation of LIM family members and their co-factors in the adult CA3 subfield
The CA3 subfield has pyramidal neurons, which receive input from the dentate granule cells. They display profound alterations in dendritic structure and branching in response to seizure. In our experiments using kainate-induced seizure, Lhx1 mRNA increased (20%, p = 0.014) in the adult CA3. In contrast, Lhx2 and Lhx9 levels decreased (30%, p = 0.028; 35%, p = 0.044 respectively; Figure 2, Figure 4a). Levels of both Lmo3 and Lmo4 were reduced (40%, p = 0.007; 25%, p = 0.002 respectively). The levels of the cofactor Clim1a also decreased (15%, p = 0.047) whereas Clim2 levels remained unaltered in the adult CA3 (Figure 2, Figure 4b).
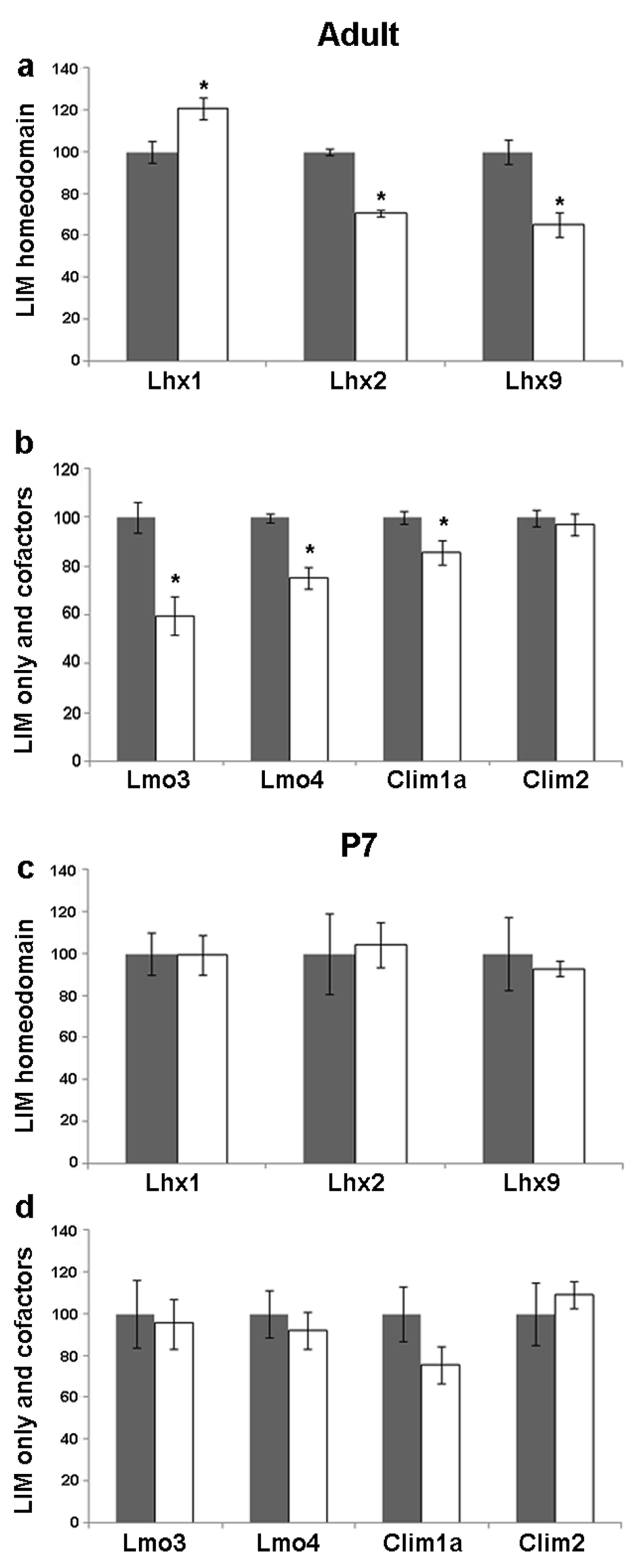
Figure 4. Kainate-induced regulation of LIM genes and co-factors in CA3 of adult and P7 rats.
Quantitative densitometric analysis of the adult CA3 region following kainate administration in adult (a,b) and P7 (c,d) rats. Grey bars are controls, white bars are kainate treated animals. Results are expressed as mean ± SEM percentage of control for mRNA expression (*p < 0.05, unpaired Student’s t test).
Seizure induced regulation of LIM family members and their co-factors in the adult CA1 subfield
The CA1 pyramidal neurons receive input from the CA3 neurons. They displayed altered dendritic shape and density and also axon sprouting as a result of seizure45. In the CA1 field, Lhx1 mRNA increased (20%, p = 0.02), whereas Lhx2 levels decreased (19%, p = 0.024) but, there was no change in Lhx9 mRNA levels in the adult CA1 upon kainate-induced seizure (Figure 2, Figure 5a). Lmo3 mRNA levels decreased (41%, p = 0.0018) whereas Lmo4, Clim1a, and Clim2 levels remained unchanged (Figure 2, Figure 5b).
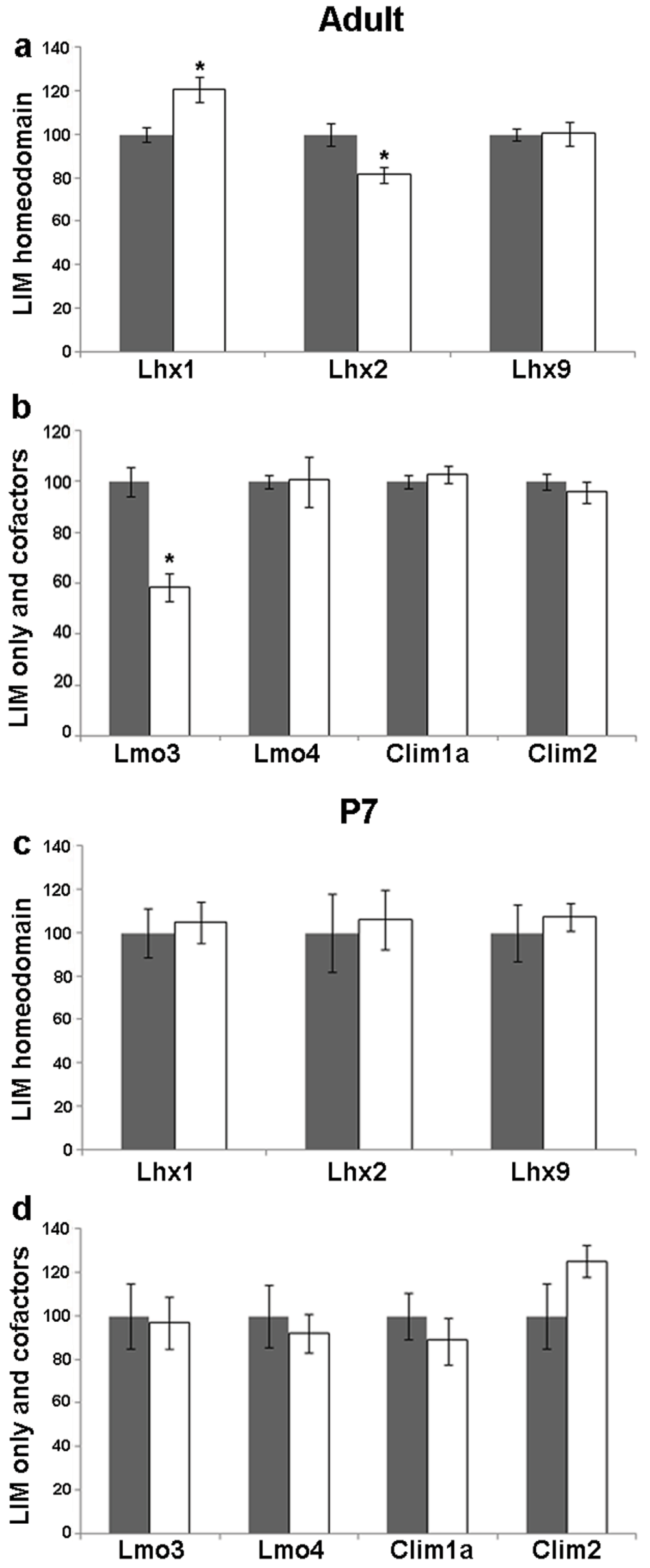
Figure 5. Kainate-induced regulation of LIM genes and co-factors in CA1 of adult and P7 rats.
Quantitative densitometric analysis of the adult CA1 region following kainate administration in adult (a,b) and P7 (c,d) rats. Grey bars are controls, white bars are kainate treated animals. Results are expressed as mean ± SEM percentage of control for mRNA expression (*p < 0.05, unpaired Student’s t test).
A summary of seizure induced regulation of LIM family members and their co-factors across all hippocampal fields
Table 1 summarizes the data such that seizure-induced regulation can be compared within a particular field as well as for a particular gene across all fields. For example, upon kainate induced seizure, Lhx1 mRNA shows a significant increase over very low baseline expression in all the hippocampal fields in response to seizure. In contrast, Lhx2 and Lmo3 show a significant decrease in all hippocampal fields. Interestingly, Lhx9 and Clim1a show a significant decrease in CA3 and DG, but not in CA1. Lmo4 transcript levels increase in the DG, decrease in the CA3 and show no change in the CA1. This correlates with the fact that the DG and CA3 undergo a more drastic structural reorganization in response to seizure46,47. Clim2 shows no alteration suggesting it may not have any additional roles in kainate-induced plasticity, but continues to be available to LIM-HD transcription factors at the same levels.
Table 1. Summary of seizure evoked regulation of LIM genes and co-factors across the hippocampal fields.
| LIM gene | DG | CA3 | CA1 |
|---|
| Adult | P7 | Adult | P7 | Adult | P7 |
|---|
|
Lhx1
| ↑ | _ | ↑ | _ | ↑ | _ |
|
Lhx2
| ↓ | _ | ↓ | _ | ↓ | _ |
|
Lhx9
| ↓ | _ | ↓ | _ | _ | _ |
|
Lmo3
| ↓ | _ | ↓ | _ | ↓ | _ |
|
Lmo4
| ↑ | _ | ↓ | _ | _ | _ |
|
Clim1a
| ↓ | _ | ↓ | _ | _ | _ |
|
Clim2
| _ | _ | _ | _ | _ | _ |
Seizures do not affect LIM-gene expression in the postnatal hippocampus
Seizure evoked structural plasticity differs between the postnatal and adult hippocampus in its extent as well as the type of changes seen. Although postnatal kainate treatment evokes powerful seizures, the immature brain is relatively resistant to seizure-evoked structural remodeling. For example, mossy fiber sprouting is absent or delayed48–50, and DG neurogenesis is unaltered or biphasically regulated with an initial decline and a delayed increase51–53 in response to seizure in the postnatal hippocampus. We asked whether the postnatal hippocampus differs from the adult hippocampus in kainic acid induced regulation of LIM genes and co-factors. We administered kainic acid to rat pups on postnatal day P7 and analyzed changes in transcript levels of several LIM genes 6 hours later. In striking contrast to the changes observed in the adult brain, the postnatal hippocampus appears refractory to regulation of the LIM-HD family following kainate evoked seizures (Figure 2, Figure 3c, d, Figure 4c, d, Figure 5c, d).
In summary, the LIM gene family and its co-factors display distinct and highly field-specific regulation in response to kainate induced seizure in the adult, but not in the postnatal hippocampus.
Discussion
Differential regulation of LIM-gene expression in response to seizures
Seizures can lead to different forms of hippocampal plasticity, which include axonal/dendritic remodeling and neurogenesis. Chemical-induced seizures like the kainic acid (kainate) treatment are used as models for epilepsy and have been shown to increase neurogenesis in the adult DG28 and extensive mossy fiber sprouting where mossy fibers aberrantly synapse onto dentate granule cells instead of CA3 pyramidal neurons41,44. Kainic acid administration causes animals to display motor signs including convulsions. In our experiments, we observed changes in the transcript levels 6 hours post kainic acid administration, after the animals displayed all the characteristic physical stages of seizures. In future experiments it would be interesting to examine whether any LIM gene transcript regulation occurs in a shorter time window post kainic acid administration, prior to the physical manifestation of seizure by the animal.
Transcription factors important for brain development are also known to regulate structural changes and reorganization in the adult brain, one example being members of the basic Helix-Loop-Helix (bHLH) family25,26. Members of the LIM-HD family of transcription factors are necessary for different aspects of the development of the hippocampus11,18,19, a structure that is vulnerable to changes in response to activity. LIM genes are differentially expressed in both the postnatal and adult hippocampus, suggesting that there might be a role for these genes in postnatal circuit development and adult reorganization16. We therefore hypothesized that the LIM-HD family members are differentially regulated in response to activity. Indeed, from our analysis of radioactive in-situ hybridization, we find that each hippocampal field displays differential expression and post-seizure regulation of different LIM genes. LIM-only genes Lmo1, 2 and 3 were previously shown to be regulated in response to kainate-induced seizures in the adult hippocampus36. We report that Lmo4 is also regulated by kainate-induced seizures throughout the hippocampus. We also discovered that LIM-HD genes Lhx1, Lhx2, Lhx9 and cofactor Clim1a are differentially regulated in response to seizures in a field-specific manner. Furthermore, we show that this differential regulation of LIM genes is restricted to adult animals and when we administered kainic acid to postnatal pups, no such regulation was observed. This is intriguing because these results highlight that a developing system such as the hippocampal circuitry in the early postnatal brain is relatively resistant to seizure-induced structural remodelling and plasticity32. For example, in the adult, seizure induces an increase in DG neurogenesis whereas in early postnatal stages, it is either decreased or unchanged34. Our results raise the intriguing possibility that such differences in molecular regulation of transcription factors may underlie the differing nature of cellular changes evoked by seizures in the postnatal versus adult brain.
Structural changes in the hippocampus
Seizure leads to an increase in neuronal activity thereby inducing the transcription of several immediate early genes (IEGs). The IEGs are hypothesized to be involved in seizure-induced structural remodelling54. The LIM family of transcription factors could be part of effector cascades downstream of these IEGs, which may eventually lead to the structural changes seen in different hippocampal subfields. CREB, a well-known activity regulated transcription factor, has been shown to interact in the same transcriptional complex as Lmo4 in response to activity55. It is also interesting to note that well known seizure-responsive IEGs in the adult hippocampus, such as the AP-1 complex, are not regulated by postnatal seizures56. This further supports the idea that distinct molecular changes evoked by postnatal versus adult seizures may contribute to the age-dependent differences in seizure-evoked plasticity.
Distinct structural changes occur in response to seizure in different subfields of the hippocampus. On seizure induction, DG shows an increase in the granule cell neurogenesis46, enhanced integration of granule cells into the neurocircuitry, a profound increase in mossy fiber sprouting by these neurons and formation of recurrent synapses57–59. The CA3 and CA1 pyramidal neurons show a loss of dendritic spine and dendritic branches47 post seizure. Some axon sprouting is also seen in CA1 neurons45,60. LIM genes may bring about activity induced structural changes in the hippocampus. They are known to regulate neurite outgrowth24. Some LIM-HD genes also control key axon guidance molecules such as Eph/ephrins61, which affect mossy fiber sprouting in the DG62. Lhx1 is known to regulate the transcription of Eph/ephrins in a subset of motor neurons61. Our results show increased Lhx1 mRNA levels in the DG in response to seizure that could lead to increased Eph/ephrin levels therefore contributing to mossy fiber sprouting. Lhx2 represses Robo1 and 2 expression in the thalamus during thalamocortical pathfinding22 and so down regulation of Lhx2 mRNA in response to seizure could be important for mossy fiber sprouting via upregulation of the Robo receptors. Lmo4 has been shown to confer a neuroprotective role in response to hypoxia63. Interestingly, we find an increase in the Lmo4 mRNA after kainate treatment, which could lead to neuronal survival in response to seizure.
Our study provides new evidence of seizure mediated regulation of LIM-HD transcription factors. We show that this regulation is age-dependent and field specific. Future experiments will aim at testing whether LIM genes are necessary for mediating seizure induced structural alterations. Examining the effect of kainic acid treatment on structural changes such as DG neurogenesis in LIM gene loss-of-function mutants will begin to address this issue. In addition, determining the interactions of LIM gene family proteins with other factors known to mediate structural changes such as the bHLH family members25,26 will open avenues for the mechanistic understanding of this process. These results therefore provide impetus for future studies to explore the role of the LIM-HD transcription factors, LIM only genes, and their cofactors in activity-dependent reorganization and plasticity in the mature nervous system.
Materials and methods
Animals and treatment paradigm
Sprague-Dawley rats were bred in the Tata Institute of Fundamental Research (TIFR) Animal house, maintained under normal 12-hour light/dark cycle and were provided with food and water ad libitum. A total of 84 adults and 101 pups (P7) were used. Adults were between 2–3 months old and weighed between 200–250 grams. All animal procedures were performed in accordance with the NIH guidelines for use and maintenance of animals and were approved by the TIFR Institutional Animal Ethics committee. The male rats were sexed at P21 and were used for experiments when they reached adulthood. Postnatal pups of both sexes were used for experiments at P7. All animals were grouped based on their treatment with either saline (control group; n = 44 adults; n = 47 P7 pups) or with 10mg/kg kainic acid (Sigma, USA; n = 40 adults; n = 54 P7 pups) administered intraperitoneally and were housed isolated for 6 hours after the treatment. The kainic acid treated group was observed every 30 minutes across the 6 hours and displayed all the characteristic stages of seizures. The animals displayed facial clonus (Racine Stage 1) to front and hindlimb clonus and continuous falling down (Racine Stage 5).
Animals were decapitated using a guillotine 6 hours after treatment and the brains were immediately frozen on dry ice and stored at -70°C. Coronal sections (14µm) were generated on the cryostat and mounted onto Probe-plus RNase free slides (Electron Microscopy Sciences, USA). Slides were then treated with 4% paraformaldehyde (PFA; Merck Chemicals), washed in 1X phosphate-buffered saline, acetylated with acetic acid (Qualigens Fine Chemicals) in 0.1M triethanolamine (Sigma-Aldrich), rinsed in 2X sodium saline citrate (SSC), pH 4.5 and then dehydrated through grades (30%, 70% and 100% in double distilled water) of ethanol (Commercial Alcohols, Ontario, Canada) prior to storage at -70°C.
mRNA in-situ hybridization
The in-situ hybridization for DIG-labeled probes was carried out as described previously (Bulchand et al., 2003)16. Plasmid DNAs encoding different LIM genes and co-factors were linearized by restriction digestion to provide template for making DIG-labeled RNA probe16. Briefly, the slides were incubated in hybridization buffer (50% formamide, 5X SSC and 1% SDS) containing DIG-labeled riboprobes (Roche) for 16 hours at 70°C followed by post-hybridization washes using Solution X (50% formamide, 2X SSC and 1% SDS), 2X SSC and 0.2X SSC.
Radioactive in-situ hybridization was carried out as described previously64. Briefly, the slides were incubated in the hybridization buffer (50% formamide, 0.6M sodium chloride, 10mM Tris pH 7.4, 1X Denhardts solution, 10mM dithiotheritol (DTT), 250µg/ml yeast tRNA, 50µg/ml Salmon sperm DNA, 10% Dextran sulphate) containing35 S-UTP labeled riboprobes (Amersham, Buckinghamshire, UK) at a concentration of 106cpm/250µl for 20–24 hours at 60°C. Post-hybridization, the slides were washed with 2XSSC, treated with RNase A (20µg/ml for 30 minutes at 37°C; USB Corporation, Cleveland, Ohio), 0.5X SSC for 30 minutes at 60°C, 0.1X SSC for 20 minutes and then rinsed in double distilled water. Slides were air dried and exposed to Biomax film (Kodak) for 3–6 weeks. To confirm the specificity of the signal observed with antisense riboprobes, controls used were sense riboprobes or RNase treatment (40µg/ml at 37°C for 30 minutes) prior to hybridization.
Quantitation and data analysis
Densitometric analysis of LIM gene transcript levels was performed using the Macintosh-based Scion Imaging software (Scion, Frederick, Maryland, USA). Sections were observed directly on the monitor using a Sony 3 CCD color video camera (Model DXC-390P).14C standards were used for calibration to correct for non-linearity. An equivalent area was outlined for each of the hippocampal subfields and optical density measurements from both hemispheres of 3–4 individual sections from each animal were analysed to calculate the mean value. Results were subjected to statistical Student’s t-test. Significance was determined at p < 0.05 using GraphPad inSTAT (version 3.05, LaJolla, California, USA). The following numbers of animals were used for each condition: Control adults, n = 7 (Clim1a), 9 (Clim2), 5 (Lhx1), 5 (Lhx2), 5 (Lhx9), 4 (Lmo3), 9 (Lmo4). Kainate treated adults, n = 8 (Clim1a), 8 (Clim2), 5 (Lhx1), 4 (Lhx2), 4 (Lhx9), 3 (Lmo3), 8 (Lmo4). Control pups, n = 6 (Clim1a), 8 (Clim2), 6 (Lhx1), 8 (Lhx2), 5 (Lhx9), 7 (Lmo3), 7 (Lmo4). Kainate treated pups, n = 8 (Clim1a), 8 (Clim2), 8 (Lhx1), 8 (Lhx2), 7 (Lhx9), 8 (Lmo3), 7 (Lmo4).
Author contributions
VL, LS, VV, ST conceived the project. VL, LS, DH, AS performed the experiments, analyzed the data, and helped to critically revise the paper. DH, VV, ST analyzed the data and wrote the paper.
Competing interests
No competing interests were disclosed.
Grant information
This work was supported by a Wellcome trust Senior Fellowship (056684/Z/99/Z), a Swarnajayanti Fellowship (Dept. of Science and Technology, Govt. of India) and a Lady Tata Memorial Trust to ST; a Wellcome Trust International Senior Research Fellowship (04082003114133) to VV, and a Kanwal Rekhi Career Development Award (Tata Institute of Fundamental Research Endowment Fund) to LS.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgements
We thank B. Anderson, F. Porter, Y. Nakagawa, I. Bach and E. A. Grove for gifts of plasmid DNA to make cRNA probes for in situ hybridization; SB Banerjee for assistance with the radioactive in-situ hybridization experiments. We thank Dr. Yashwantrao Mane and the TIFR Animal House staff for excellent support.
Faculty Opinions recommendedReferences
- 1.
Guillemot F:
Cell fate specification in the mammalian telencephalon.
Prog Neurobiol.
2007; 83(1): 37–52. PubMed Abstract
| Publisher Full Text
- 2.
Polleux F, Ince-Dunn G, Ghosh A:
Transcriptional regulation of vertebrate axon guidance and synapse formation.
Nat Rev Neurosci.
2007; 8(5): 331–40. PubMed Abstract
| Publisher Full Text
- 3.
Nóbrega-Pereira S, Marín O:
Transcriptional control of neuronal migration in the developing mouse brain.
Cereb Cortex.
2009; 19(Suppl 1): i107–13. PubMed Abstract
| Publisher Full Text
- 4.
Lonze BE, Ginty DD:
Function and regulation of CREB family transcription factors in the nervous system.
Neuron.
2002; 35(4): 605–23. PubMed Abstract
| Publisher Full Text
- 5.
Parrish JZ, Emoto K, Kim MD, et al.:
Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields.
Annu Rev Neurosci.
2007; 30: 399–423. PubMed Abstract
| Publisher Full Text
- 6.
West AE, Greenberg ME:
Neuronal activity-regulated gene transcription in synapse development and cognitive function.
Cold Spring Harb Perspect Biol.
2011; 3(6): a005744. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 7.
Lundgren SE, Callahan CA, Thor S, et al.:
Control of neuronal pathway selection by the Drosophila LIM homeodomain gene apterous.
Development.
1995; 121(6): 1769–73. PubMed Abstract
- 8.
Porter FD, Drago J, Xu Y, et al.:
Lhx2, a LIM homeobox gene, is required for eye forebrain, and definitive erythrocyte development.
Development.
1997; 124(15): 2935–4. PubMed Abstract
- 9.
Thor S, Andersson SG, Tomlinson A, et al.:
A LIM-homeodomain combinatorial code for motor-neuron pathway selection.
Nature.
1999; 397(6714): 76–80. PubMed Abstract
| Publisher Full Text
- 10.
Ando H, Kobayashi M, Tsubokawa T, et al.:
Lhx2 mediates the activity of Six3 in zebrafish forebrain growth.
Dev Biol.
2005; 287(2): 456–468. PubMed Abstract
| Publisher Full Text
- 11.
Mangale VS, Hirokawa KE, Satyaki PR, et al.:
Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate.
Science.
2008; 319(5861): 304–309. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 12.
Milan M, Cohen SM:
Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO.
Mol Cell.
1999; 4(2): 267–273. PubMed Abstract
| Publisher Full Text
- 13.
Thaler JP, Lee SK, Jurata LW, et al.:
LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions.
Cell.
2002; 110(2): 237–249. PubMed Abstract
| Publisher Full Text
- 14.
Milan M, Diaz-Benjumea FJ, Cohen SM:
Beadex encodes an LMO protein that regulates Apterous LIM-homeodomain activity in Drosophila wing development: a model for LMO oncogene function.
Genes Dev.
1998; 12(18): 2912–2920. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 15.
Weihe U, Milan M, Cohen SM:
Regulation of Apterous activity in Drosophila wing development.
Development.
2001; 128(22): 4615–4622. PubMed Abstract
- 16.
Bulchand S, Subramanian L, Tole S:
Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex.
Dev Dyn.
2003; 226(3): 460–469. PubMed Abstract
| Publisher Full Text
- 17.
Bulchand S, Grove EA, Porter FD, et al.:
LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem.
Mech Dev.
2001; 100(2): 165–175. PubMed Abstract
| Publisher Full Text
- 18.
Subramanian L, Sarkar A, Shetty AS, et al.:
Transcription factor Lhx2 is necessary and sufficient to suppress astrogliogenesis and promote neurogenesis in the developing hippocampus.
Proc Natl Acad Sci U S A.
2011; 108(27): E265–74. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 19.
Zhao Y, Sheng HZ, Amini R, et al.:
Control of hippocampal morphogenesis and neuronal differentiation by the LIM homeobox gene Lhx5.
Science.
1999; 284(5417): 1155–8. PubMed Abstract
| Publisher Full Text
- 20.
Yan CH, Levesque M, Claxton S, et al.:
Lmx1a and lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors.
J Neurosci.
2011; 31(35): 12413–25. PubMed Abstract
| Publisher Full Text
- 21.
Lakhina V, Falnikar A, Bhatnagar L, et al.:
Early thalamocortical tract guidance and topographic sorting of thalamic projections requires LIM-homeodomain gene Lhx2.
Dev Biol.
2007; 306(2): 703–13. PubMed Abstract
| Publisher Full Text
- 22.
Marcos-Mondéjar P, Peregrín S, Li JY, et al.:
The lhx2 transcription factor controls thalamocortical axonal guidance by specific regulation of robo1 and robo2 receptors.
J Neurosci.
2012; 32(13): 4372–85. PubMed Abstract
| Publisher Full Text
- 23.
Hobert O, Tessmar K, Ruvkun G:
The Caenorhabditis elegans lim-6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons.
Development.
1999; 126(7): 1547–1562. PubMed Abstract
- 24.
Manetopoulos C, Hansson A, Karlsson J, et al.:
The LIM-only protein LMO4 modulates the transcriptional activity of HEN1.
Biochem Biophys Res Commun.
2003; 307(4): 891–9. PubMed Abstract
| Publisher Full Text
- 25.
Elliott RC, Khademi S, Pleasure SJ, et al.:
Differential regulation of basic helix-loop-helix mRNAs in the dentate gyrus following status epilepticus.
Neuroscience.
2001; 106(1): 79–88. PubMed Abstract
| Publisher Full Text
- 26.
Elliott RC, Miles MF, Lowenstein DH:
Overlapping microarray profiles of dentate gyrus gene expression during development- and epilepsy-associated neurogenesis and axon outgrowth.
J Neurosci.
2003; 23(6): 2218–27. PubMed Abstract
- 27.
Takasu MA, Dalva MB, Zigmond RE, et al.:
Modulation of NMDA receptor-dependent calcium influx and gene expression through EphB receptors.
Science.
2002; 295(5554): 491–5. PubMed Abstract
| Publisher Full Text
- 28.
Parent JM, Lowenstein DH:
Seizure-induced neurogenesis: are more new neurons good for an adult brain?
Prog Brain Res.
2002; 135: 121–131. PubMed Abstract
| Publisher Full Text
- 29.
Nadler JV:
The recurrent mossy fiber pathway of the epileptic brain.
Neurochem Res.
2003; 28(11): 1649–58. PubMed Abstract
| Publisher Full Text
- 30.
Overstreet-Wadiche LS, Bromberg DA, Bensen AL, et al.:
Seizures accelerate functional integration of adult-generated granule cells.
J Neurosci.
2006; 26(15): 4095–103. PubMed Abstract
| Publisher Full Text
- 31.
Scharfman HE, McCloskey DP:
Postnatal neurogenesis as a therapeutic target in temporal lobe epilepsy.
Epilepsy Res.
2009; 85(2–3): 150–61. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 32.
Sperber EF, Haas KZ, Stanton PK, et al.:
Resistance of the immature hippocampus to seizure-induced synaptic reorganization.
Brain Res Dev Brain Res.
1991; 60(1): 88–93. PubMed Abstract
| Publisher Full Text
- 33.
Lynch M, Sayin U, Bownds J, et al.:
Long-term consequences of early postnatal seizures on hippocampal learning and plasticity.
Eur J Neurosci.
2000; 12(7): 2252–64. PubMed Abstract
| Publisher Full Text
- 34.
Porter BE:
Neurogenesis and epilepsy in the developing brain.
Epilepsia.
2008; 49(Suppl 5): 50–4. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 35.
Alberini CM:
Transcription factors in long-term memory and synaptic plasticity.
Physiol Rev.
2009; 89(1): 121–45. PubMed Abstract
| Publisher Full Text
- 36.
Hinks GL, Shah B, French SJ, et al.:
Expression of LIM protein genes Lmo1, Lmo2, and Lmo3 in adult mouse hippocampus and other forebrain regions: differential regulation by seizure activity.
J Neurosci.
1997; 17(14): 5549–59. PubMed Abstract
- 37.
Altman J, Das GD:
Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats.
J Comp Neurol.
1965; 124(3): 319–35. PubMed Abstract
| Publisher Full Text
- 38.
Guéneau G, Privat A, Drouet J, et al.:
Subgranular zone of the dentate gyrus of young rabbits as a secondary matrix. A high-resolution autoradiographic study.
Dev Neurosci.
1982; 5(4): 345–58. PubMed Abstract
| Publisher Full Text
- 39.
Eckenhoff MF, Rakic P:
Nature and fate of proliferative cells in the hippocampal dentate gyrus during the life span of the rhesus monkey.
J Neurosci.
1988; 8(8): 2729–47. PubMed Abstract
- 40.
Gray WP, Sundstrom LE:
Kainic acid increases the proliferation of granule cell progenitors in the dentate gyrus of the adult rat.
Brain Res.
1998; 790(1–2): 52–9. PubMed Abstract
| Publisher Full Text
- 41.
Wenzel HJ, Woolley CS, Robbins CA, et al.:
Kainic acid-induced mossy fiber sprouting and synapse formation in the dentate gyrus of rats.
Hippocampus.
2000; 10(3): 244–60. PubMed Abstract
| Publisher Full Text
- 42.
Lein ES, Zhao X, Gage FH:
Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization.
J Neurosci.
2004; 24(15): 3879–89. PubMed Abstract
| Publisher Full Text
- 43.
Dong H, Csernansky CA, Goico B, et al.:
Hippocampal neurogenesis follows kainic acid-induced apoptosis in neonatal rats.
J Neurosci.
2003; 23(5): 1742–9. PubMed Abstract
- 44.
Okazaki MM, Evenson DA, Nadler JV:
Hippocampal mossy fiber sprouting and synapse formation after status epilepticus in rats: visualization after retrograde transport of biocytin.
J Comp Neurol.
1995; 352(4): 515–34. PubMed Abstract
| Publisher Full Text
- 45.
Smith BN, Dudek FE:
Short- and long-term changes in CA1 network excitability after kainate treatment in rats.
J Neurophysiol.
2001; 85(1): 1–9. PubMed Abstract
- 46.
Parent JM, Yu TW, Leibowitz RT, et al.:
Dentate granule cell neurogenesis is increased by seizures and contributes to aberrant network reorganization in the adult rat hippocampus.
J Neurosci.
1997; 17(10): 3727–38. PubMed Abstract
- 47.
Jiang M, Lee CL, Smith KL, et al.:
Spine loss and other persistent alterations of hippocampal pyramidal cell dendrites in a model of early-onset epilepsy.
J Neurosci.
1998; 18(20): 8356–68. PubMed Abstract
- 48.
Ribak CE, Navetta MS:
An immature mossy fiber innervation of hilar neurons may explain their resistance to kainate-induced cell death in 15–day-old rats.
Brain Res Dev Brain Res.
1994; 79(1): 47–62. PubMed Abstract
| Publisher Full Text
- 49.
Cornejo BJ, Mesches MH, Coultrap S, et al.:
A single episode of neonatal seizures permanently alters glutamatergic synapses.
Ann Neurol.
2007; 61(5): 411–26. PubMed Abstract
| Publisher Full Text
- 50.
Cross DJ, Cavazos JE:
Synaptic reorganization in subiculum and CA3 after early-life status epilepticus in the kainic acid rat model.
Epilepsy Res.
2007; 73(2): 156–65. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 51.
Gray WP, May K, Sundstrom LE:
Seizure induced dentate neurogenesis does not diminish with age in rats.
Neurosci Lett.
2002; 330(3): 235–8. PubMed Abstract
| Publisher Full Text
- 52.
Bender RA, Dubé C, Gonzalez-Vega R, et al.:
Mossy fiber plasticity and enhanced hippocampal excitability, without hippocampal cell loss or altered neurogenesis, in an animal model of prolonged febrile seizures.
Hippocampus.
2003; 13(3): 399–412. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 53.
Liu H, Kaur J, Dashtipour K, et al.:
Suppression of hippocampal neurogenesis is associated with developmental stage, number of perinatal seizure episodes, and glucocorticosteroid level.
Exp Neurol.
2003; 184(1):196–213. PubMed Abstract
| Publisher Full Text
- 54.
Watanabe Y, Johnson RS, Butler LS, et al.:
Null mutation of c-fos impairs structural and functional plasticities in the kindling model of epilepsy.
J Neurosci.
1996; 16(12): 3827–36. PubMed Abstract
- 55.
Kashani AH, Qiu Z, Jurata L, et al.:
Calcium activation of the LMO4 transcription complex and its role in the patterning of thalamocortical connections.
J Neurosci.
2006; 26(32): 8398–408. PubMed Abstract
| Publisher Full Text
- 56.
Pennypacker KR, McMillian MK, Douglass J, et al.:
Ontogeny of kainate-induced gene expression in rat hippocampus.
J Neurochem.
1994; 62(2): 438–44. PubMed Abstract
| Publisher Full Text
- 57.
Tauck DL, Nadler JV:
Evidence of functional mossy fiber sprouting in hippocampal formation of kainic acid-treated rats.
J Neurosci.
1985; 5(4): 1016–22. PubMed Abstract
- 58.
Represa A, Jorquera I, Le Gal La Salle G, et al.:
Epilepsy induced collateral sprouting of hippocampal mossy fibers: does it induce the development of ectopic synapses with granule cell dendrites?
Hippocampus.
1993; 3(3): 257–68. PubMed Abstract
| Publisher Full Text
- 59.
Lynch M, Sutula T:
Recurrent excitatory connectivity in the dentate gyrus of kindled and kainic acid-treated rats.
J Neurophysiol.
2000; 83(2): 693–704. PubMed Abstract
- 60.
Perez Y, Morin F, Beaulieu C, et al.:
Axonal sprouting of CA1 pyramidal cells in hyperexcitable hippocampal slices of kainate-treated rats.
Eur J Neurosci.
1996; 8(4): 736–748. PubMed Abstract
| Publisher Full Text
- 61.
Lee SK, Pfaff SL:
Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors.
Neuron.
2003; 38(5): 731–45. PubMed Abstract
| Publisher Full Text
- 62.
Xu B, Li S, Brown A, et al.:
EphA/ephrin-A interactions regulate epileptogenesis and activity-dependent axonal sprouting in adult rats.
Mol Cell Neurosci.
2003; 24(4): 984–99. PubMed Abstract
| Publisher Full Text
- 63.
Chen HH, Schock SC, Xu J, et al.:
Extracellular ATP-dependent upregulation of the transcription cofactor LMO4 promotes neuron survival from hypoxia.
Exp Cell Res.
2007; 313(14): 3106–16. PubMed Abstract
| Publisher Full Text
- 64.
Nair A, Vadodaria KC, Banerjee SB, et al.:
Stressor-specific regulation of distinct brain-derived neurotrophic factor transcripts and cyclic AMP response element-binding protein expression in the postnatal and adult rat hippocampus.
Neuropsychopharmacology.
2007; 32(7): 1504–19. PubMed Abstract
| Publisher Full Text





Comments on this article Comments (0)