Keywords
SV2; nucleotide binding; ATP hydrolysis
SV2; nucleotide binding; ATP hydrolysis
Synaptic vesicle protein 2 (SV2) is a membrane protein expressed exclusively in neurons and endocrine cells where it is localized to vesicles that undergo regulated secretion1. Most mammals have three SV2 genes that encode three related proteins termed SV2A, SV2B, and SV2C2,3. Of these, SV2A is the most broadly expressed, and is present in nearly all neurons, whereas SV2C demonstrates the most restricted expression2,4,5. SV2 is essential for normal neurotransmission, as evidenced by epileptic seizures and premature death in SV2A or SV2A/B knockout mice6,7. Electrophysiological studies assessing neurotransmitter release in SV2 mutants indicate that SV2 acts as a positive modulator of calcium-dependent exocytosis8–10. Neuron or endocrine cells lacking SV2 contain fewer vesicles competent for Ca2+-triggered secretion.
Despite our understanding of the physiological role of SV2, its molecular action remains unknown. We previously reported that SV2 binds adenine nucleotides including ATP11. Analysis of ATP transport by vesicles isolated from wild-type and SV2 mutants revealed that SV2 is not an ATP transporter, nor does it affect ATP transport11. Thus it is not clear how ATP binding relates to SV2 function. One untested possibility is that SV2 is an enzyme that hydrolyzes ATP. To address this, we investigated whether SV2 has ATPase activity. Our results show that recombinant SV2A and SV2B do not hydrolyze ATP. Thus, binding to adenine nucleotides likely modulates other actions of SV2.
HEK293T cell (ATCC, Manassas, VA) culture and transfection with Lipofectamine™ 2000 reagent (Invitrogen, Grand Island, NY) were performed following the manufacturer's protocol11. To transfect one 15 cm plate of cells, 48 µg of plasmid DNA and 96 µl of Lipofectamine™ 2000 reagent were mixed with 3 ml of serum and antibiotic-free minimum essential media (MEM) (Invitrogen, Grand Island, NY). After incubation at room temperature for 5 min, the DNA and Lipofectamine™ 2000 reagent mixture was combined and allowed to sit for 20 min at room temperature. The transfection mixture was then added drop-wise to cells cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum. Cells were maintained at 37°C in a humidified incubator containing 5% CO2. After 48 h, medium was removed, cells were harvested, and rinsed with ice-cold phosphate-buffered saline (PBS) two times. Cell pellets were stored at -80°C until use. Rat SV2A and SV2B were expressed with a C-terminal FLAG epitope (DYKDDDK) in the mammalian expression vector pIRES2-EGFP (Clontech, Mountain View, CA).
Recombinant SV2A and SV2B -FLAG fusion proteins were purified using anti-FLAG M2 affinity agarose beads (Sigma; catalog number: A2220, mouse IgG1 monoclonal antibody) as described previously11. Briefly, transfected cells were harvested and washed with ice-cold phosphate-buffered saline (PBS). Cell extracts were made by adding extraction buffer (150 mM potassium acetate (J.T. Baker, Phillipsburg, NJ), 10 mM HEPES-KOH (pH 7.4) (Sigma, St. Louis, MO), 1% Triton X-100 (Roche Applied Science, Indianapolis, IN), with 1× fresh protease inhibitor mixture added before each use (Roche Applied Science, Indianapolis, IN)) and incubated at 4°C for 1 h. After extraction, insoluble material was removed by centrifugation (Beckman, Indianapolis IN) at 19,000 × g for 20 min. The resulting extract was mixed with pre-equilibrated anti-FLAG M2 agarose beads (Sigma, St. Louis, MO) and incubated with agitation at 4°C for 3–4 h. The beads were washed four times with 20 volumes of extraction buffer. FLAG fusion protein was eluted from the beads with 3× FLAG peptide (Sigma, St. Louis, MO) in buffer containing 150 mM potassium acetate, 10 mM HEPES-KOH (pH 7.4) and 0.5% Triton X-100. Final preparations were checked by silver staining (Thermo, Waltham, MA) of SDS polyacrylamide gels (Bio-Rad, Hercules, CA) and immunoblot with anti-SV2 (mouse monoclonal antibody against an epitope located on the N-terminus of SV2)12 or anti-FLAG antibodies (Sigma, catalog number: F3165, mouse IgG1 monoclonal antibody). Protein concentration was determined by comparison to bovine serum albumin (BSA) (Fisher, Pittsburgh, PA) standards in PAGE gels. This method was used because the 3× FLAG peptide in the eluate will affect solution-based protein assays. BSA standards, ranging from 0.5 to 4 µg were loaded on the same gel along with purified SV2-FLAG fusion proteins. Gels stained with coomassie blue were scanned with a Kodak Image Station 440CF (Kodak, Rochester, NY). Net intensities of protein bands were determined by Kodak Molecular Imaging software (Kodak, Rochester, NY) and protein concentration of recombinant SV2-FLAG was calculated according to BSA standards.
ATP hydrolysis reactions were conducted at 25°C in a buffer consisting of 150 mM potassium acetate, 10 mM HEPES (pH 7.4), 2 mM ATP, 3 MgCl2, 0.1% Triton X-100, 1 mM DTT and 10–20 µg/ml purified recombinant SV2-FLAG protein. The ATP hydrolysis reaction was set up on ice and recombinant SV2-FLAG protein was added last. Reaction mixtures were prepared on ice, and an aliquot of reaction was immediately withdrawn as the 0°C, 0 min sample. The reaction was shifted to 25°C, and at designated intervals (1 min, 2 min, 5 min and 10 min), aliquots were removed from the reaction for analysis by the malachite green assay described below. Duplicates were assayed for each time point and the average value of the duplicates was used for quantification.
Commercially available calf intestinal alkaline phosphatase (CIP) (New England Biolabs, Ipswich, MA) was used as a positive control for the ATP hydrolysis reaction. As a negative control, an aliquot of CIP was heated at 95°C for 20 min to inactivate the enzyme.
To prepare the malachite green solution, one volume of 4.2% ammonium molybdate (Sigma, St. Louis, MO) in 4 M HCl was added to 3 volumes of 0.045% malachite green (Sigma, St. Louis, MO). The solution was stirred at room temperature for a minimum of 30 min before being filtered through a 0.22 µm membrane (Millipore, Billerica, MA). Malachite green solutions were kept at 4°C in the dark. Before each assay, 0.01% Tween-20 was added to the malachite green solution.
50 µl aliquots of ATP hydrolysis reaction were added to 950 µl of malachite green solution and color was allowed to develop for 20 min at room temperature. Absorbance at 620 nm was measured in a cuvette with the malachite green solution as a blank in a SmartSpec3000 spectrometer (Bio-Rad, Hercules, CA). A 0.02 mM KH2PO4 solution (Fisher, Pittsburgh, PA) was used as an inorganic phosphate standard. Phosphate release in ATP hydrolysis reactions was quantified by comparison to inorganic phosphate standards.
To determine if SV2 is an ATPase we used a fast and reproducible method for measuring inorganic free phosphate released by ATP hydrolysis in aqueous solutions13,14. The assay is based on the change of absorbance at 620 nm after free phosphate forms a colored complex with molybdate/malachite green15. Figure 1 shows a standard curve testing 0–400 pmol of KH2PO4 in the reaction. It indicates that the cuvette-based assay produced a reliable linear standard curve.
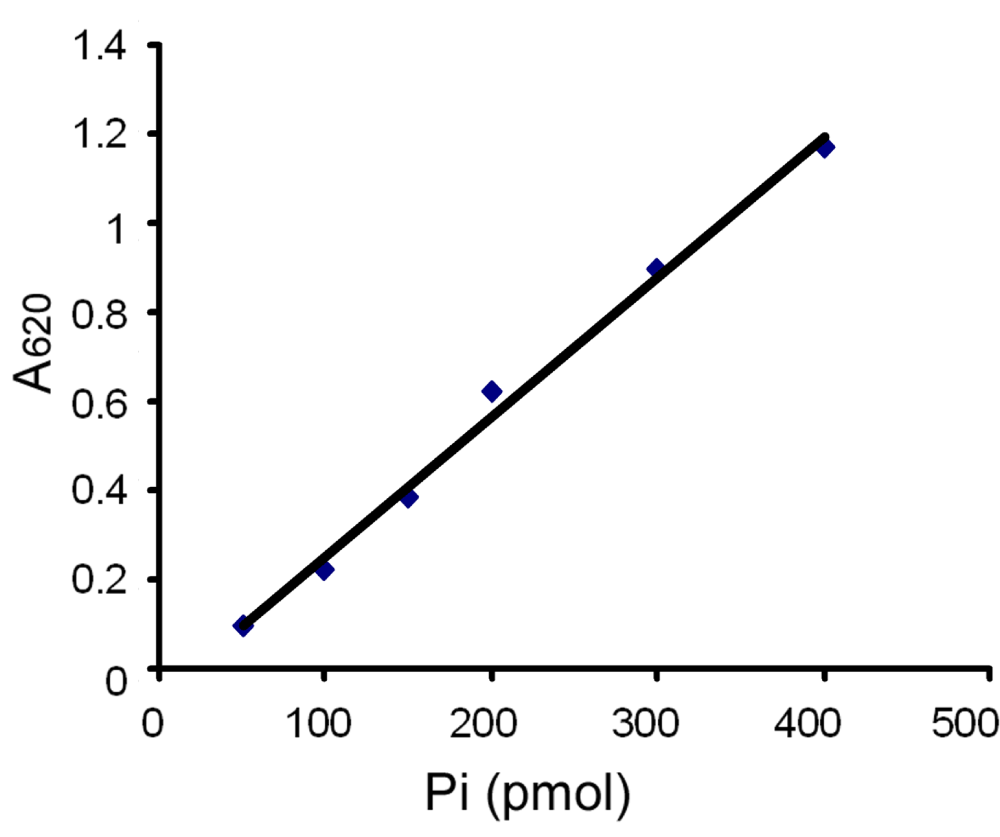
Shown is a standard curve for inorganic phosphate (Pi) in the malachite green assay. The data are representative of 5 independent experiments. Inorganic phosphate standards were prepared from KH2PO4. Each point represents the mean value of duplicate measurements. 50 µl of standard solution containing different amounts of phosphate (Pi) was mixed with 950 µl of malachite green solution and allowed to develop color at room temperature for 20 min. The absorbance of each sample was measured at 620 nm and plotted against pmol of phosphate in the reaction.
As a positive control for phosphate production, we used CIP16, a type of alkaline phosphatase that catalyzes the hydrolysis of 5´-phosphate groups from various substrates, including 5´-nucleotides, RNA and DNA. Figure 2 shows that CIP catalyzed ATP hydrolysis in a time-dependent manner. Heat-inactivated CIP produced little activity that did not increase with time. These results attest to the effectiveness of the ATP hydrolysis reaction and the malachite green assay in this study.
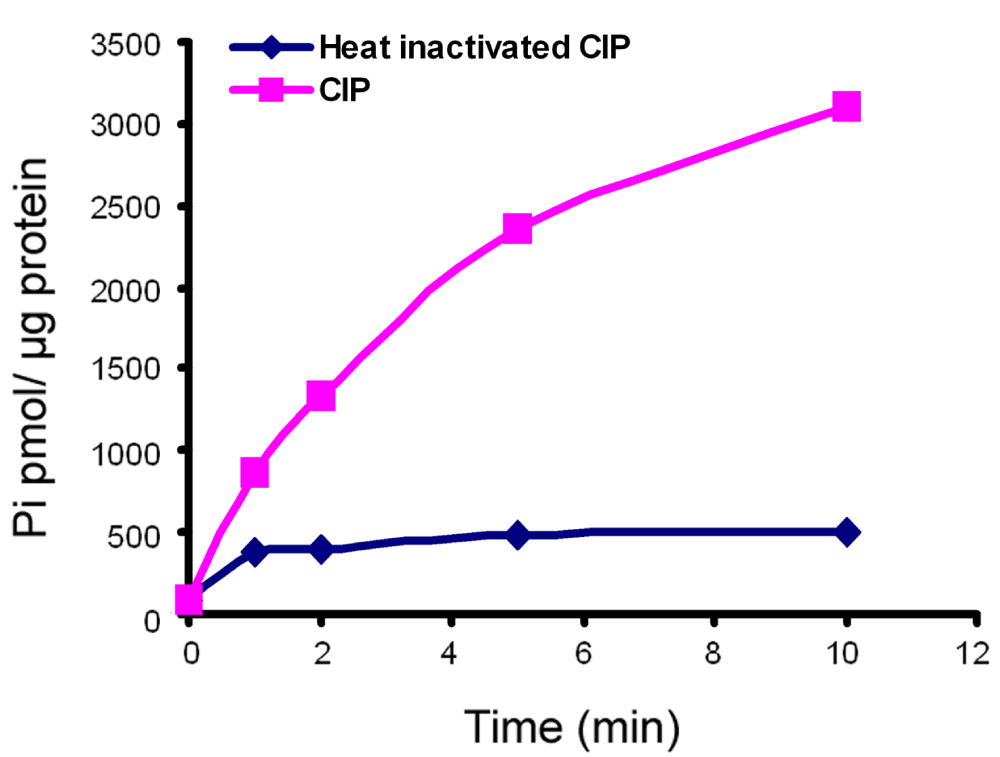
Shown is a time course of one calf intestinal alkaline phosphatase (CIP) reaction with ATP as a substrate. The data are representative of four independent experiments. The CIP reaction was carried out at 25°C. At different time points, aliquots were withdrawn and subjected to the malachite green assay as described in Figure 1. Pi in each sample was quantified by comparing to the standard curve generated for inorganic phosphate. Each point represents the mean value of duplicate measurements. CIP exhibited time-dependent ATP hydrolysis activity while heat-inactivated CIP produce much lower activity that was not time-dependent.
To test for ATPase activity in SV2, we performed the assay with purified recombinant SV2. SV2A or SV2B fused to a FLAG epitope (for affinity purification) is the same preparation that was used to characterize ATP binding activity in vitro in our previous study11. In the malachite green assay neither SV2A nor SV2B produced ATP hydrolysis activity (Figure 3).

Shown is a representative plot of ATP hydrolysis activity by recombinant SV2A (Panel A) or SV2B (Panel B). ATP hydrolysis reactions and the malachite green assay were performed as described under "Methods". The data represent two (SV2A) and three (SV2B) separate measures. For comparison, results of calf intestinal alkaline phosphatase (CIP) reactions run on the same day are included in the plots.
This result indicates that SV2 does not hydrolyze ATP. The concern that the FLAG epitope included at the carboxy terminus of recombinant SV2 used in these assays might affect activity is not likely given that SV2-EGFP fusion proteins can rescue normal neurotransmission in neurons from SV2 knockouts17. Together with previous studies reporting that SV2 is not likely to be an ATP transporter11, these findings exclude the possibility that SV2 affects ATP levels either by transport or hydrolysis. Therefore SV2 binding of ATP likely regulates its conformation or its molecular interactions, possibilities that suggest SV2 action is modulated by synaptic energy levels.
We previously show that a tyrosine-based endocytosis motif (Y46SRF) in the N-terminus of SV2 is required for its trafficking to synaptic vesicles, and that this motif mediates binding of multiple endocytosis-related proteins18. Interestingly, one of the two ATP binding sites in SV2A (a.a. 58–104) is located in the same region11. This suggests a possible link between ATP binding and SV2 endocytosis.
SV2, synaptic vesicle protein 2; CIP, calf intestinal alkaline phosphatase; ATP, adenosine-5´-triphosphate
JY and SB conceived and designed the study. JY performed the experiments and analyzed the data. JY and SB prepared the manuscript.
This work was supported by National Institutes of Mental Health Grant R01 MH 059842 to SB.
We thank Dr Alex Merz for help with the malachite green assay and Mario Rosasco for editorial review of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 09 Oct 13 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)