Introduction and aims
Idiopathic inflammatory myopathies (IIM) are chronic autoimmune disorders characterized by skeletal muscle weakness, inflammatory immune cells in muscle tissues and frequent presence of serum autoantibodies. IIM can be divided into several groups: dermatomyositis (DM), polymyositis (PM), inclusion body myositis (IBM), and immune mediated necrotizing myopathy (NM). The etiology of IIM remains unclear; however current findings indicate that epigenetic mechanisms may contribute to the disease pathogenesis1,2.
MicroRNA molecules are part of the epigenetic regulatory network. They act as negative regulators of gene expression and participate in the regulation of key biological processes such as cell growth, differentiation, proliferation or apoptosis3–6. Recent findings highlight the crucial importance of miRNAs in development, homeostasis and function of innate and adaptive immunity7–10. Aberrant expression patterns have been documented in a broad range of diseases including autoimmune disorders11.
In our study we have focused on the miR-23b type of microRNA, as it represents a novel promising autoimmunity regulator molecule with considerable therapeutic potential12. Recent research has revealed downregulation of this miRNA in resident cells present in inflammatory lesions of patients with rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE) and its role during the pathogenesis of autoimmune disease has been functionally characterized12.
The aim of our study was to analyze differential expression of miR-23b and miR-23b* in peripheral blood mononuclear cells (PBMCs) of patients with PM/DM, and to examine potential involvement of these miRNA types in the pathogenesis of IIM.
Material and methods
A cohort of 25 adult patients (age range 21–86 years, 7 males) suffering from idiopathic inflammatory myopathy and 22 healthy controls (age range 25–79 years, 5 males), both from the middle region of Czech Republic were analyzed. Diagnosis of PM or DM was determined according to the Bohan and Peter criteria13,14. In total 12 of patients suffered from PM, 13 patients were diagnosed as DM. All individuals involved in this study signed an informed consent. PBMCs of all individuals were obtained and purified by density gradient centrifugation on Ficoll in Leucosep tubes (Greiner Bio-One GmbH, Germany). 10 ml of peripheral blood of patients with myositis and healthy control subjects was collected into EDTA coated tubes under sterile conditions. The blood was added to a LeucoSep tube. The cell separation tubes were centrifuged (Jouan CR 3i; Jouan, St. Herblain, France) for 10 min at 2400 rpm without braking at room temperature. The cell suspension was collected, and the cells were washed three times in PBS (for 10 min at 1000 rpm), resuspended in 1 ml PBS and transferred into a 1,5 ml microcentrifuge tube. The microcentrifuge tube was centrifuged at 8000 rpm for 1 minute, supernatant was removed and the cell pellet was frozen using liquid nitrogen and stored at -80°C. The total RNA preparation from PBMCs was carried out using the conventional TRIZOL® reagent (Invitrogen, Carlsbad, USA) extraction procedure15. Afterwards, the RNA was treated with RQ1 RNase free DNase (Promega, USA) for 20 min at 37°C in order to remove genomic DNA contamination, purified with phenol-chloroform-isoamylalcohol (25:24:1, PENTA, Czech Republic) reextraction and precipitated by 1 volume 2-propanol and 1/10 volume 300 mM sodium acetate (pH 4.8). Then, the RNA was washed twice with 70% ethanol and dissolved in RNase-free water (Life Technologies, Carlsbad, CA, USA). The quantity of total RNA was measured with a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, USA). A microfluidic electrophoresis 2100 Bioanalyzer (Agilent Technologies, USA) was then used to quantify miRNA in absolute amounts [pg] and as a percentage of small RNA [%].
TaqMan gene specific miRNA assays were used to quantify the expression levels of mature miR-23b and miR-23b* (microRNA 000400, 002126) from Life Technologies (Carlsbad, CA, USA). Total RNA was reverse-transcribed by the TaqMan microRNA reverse transcription kit (Applied Biosystems) in a reaction mixture containing a miR-specific stem-loop reverse transcription (RT) primer. The quantification of mature miRNAs was performed using the TaqMan miRNA assay kit (Applied Biosystems) containing TaqMan primers in a universal PCR master mix without AmpErase UNG. 4 small nuclear RNAs - RNU44, RNU48, U47 and RNU6B (microRNA 001094, 001006, 001223, 001093, Applied Biosystems) were amplified as an internal control. qPCR was conducted at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The expression level of miR-23b (miR-23b*) is shown as delta Ct. The delta Ct value was normalized to the mean value of four other miRNA molecules used in this experiment as housekeeping molecules (RNU44, RNU48, RNU6B, U47). This process was used to overcome the differences in cDNA quantity between different samples. All analyses were done in technical duplicates.
Statistical significance was determined by the Student's t-test using Microsoft Excel®2007. A value of P<0.05 was considered to be statistically significant.
The whole study, including the laboratory work was performed at the Institute of Rheumatology in Prague. Collection of clinical data and blood from patients as well as the laboratory analysis of the biological material was undertaken under the regulation of the relevant local research ethics committee (Ref. Nr. 5371/2012).
Results and discussion
We have examined the differential expression of the miR-23b and miR-23b* in PBMCs of patients with PM/DM and healthy controls. The detection of expression of miR-23b and miR-23b* was performed by quantitative real-time PCR. The median expression level (delta Ct) of miR-23b in the group of IIM patients was -3.90, the expression (delta Ct) in the control group reached -4.03. The median expression level of miR-23b* was -11.69 in IIM patients and -11.68 in the controls (Figure 1). The expression level (delta Ct) of both tested miR-23b molecules did not show any significant differences between patients suffering from myositis and control cohort (Figure 1). Furthermore, we did not find any difference in miR-23b (or miR-23b*) expression between DM and PM patients. The table with raw qPCR expression data used for statistical evaluation is provided in supplementary material.
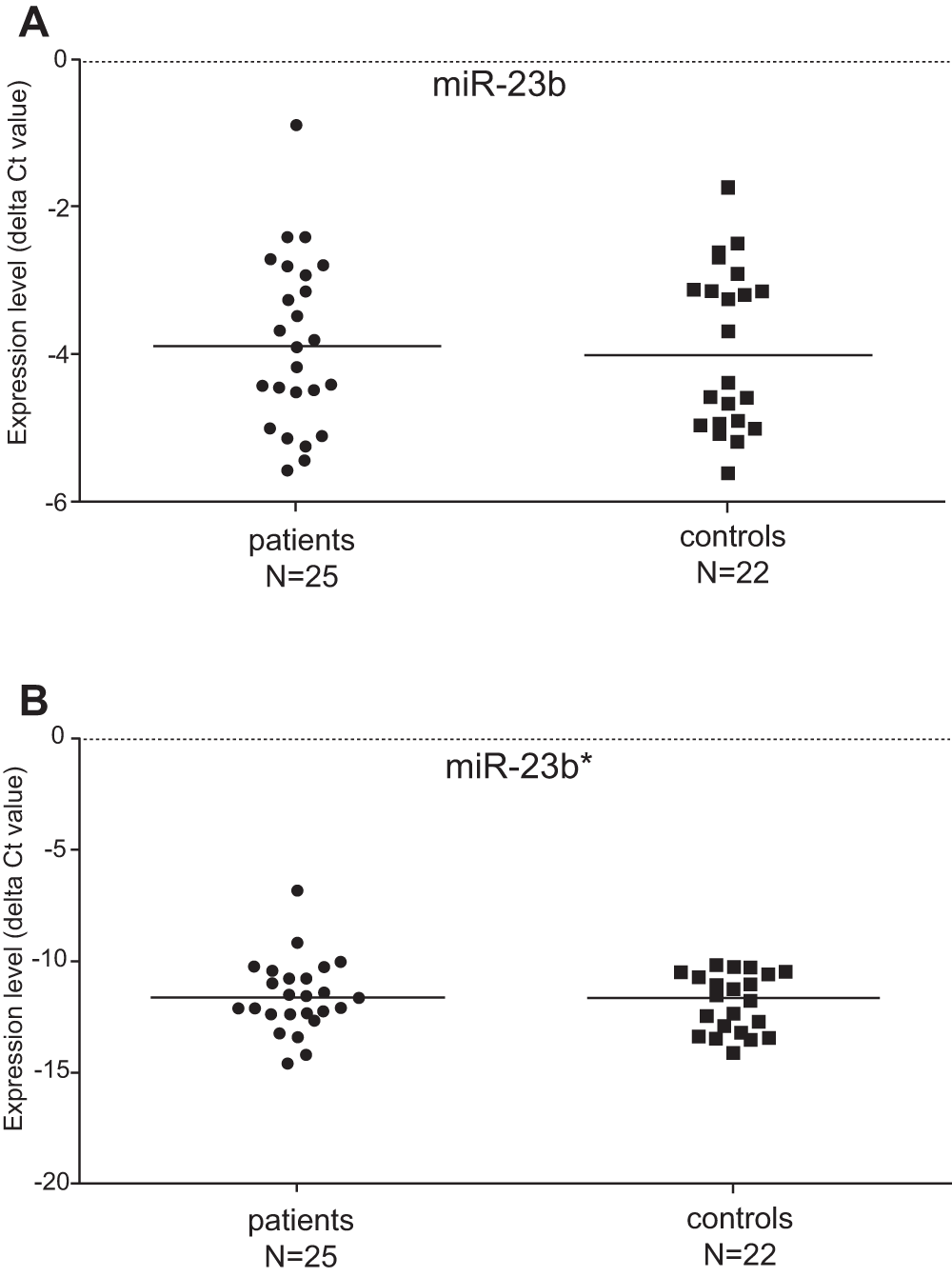
Figure 1. Regulation of the miR-23b and miR-23b* expression in patients and controls.
The expression level of miR-23b was not significantly changed in patients suffering from idiopathic inflammatory myopathy when compared with healthy controls (A). No significant difference in expression of miR-23b* was found between myositis patients and controls (B).
The role of miR-23b has been studied on inflammatory lesions of humans with lupus erythematosus or rheumatoid arthritis, as well as on corresponding tissues of murine models of lupus, rheumatoid arthritis and multiple sclerosis12. The authors Zhu et al.12 demonstrated that miR-23b targets TAB2, TAB3 and IKK-α mRNA transcripts and thereby suppresses autoimmune symptomatology by limiting the actions of the key proinflammatory cytokines IL-17, IL-1β and TNF. As it was found that the in vitro treatment of resident cell lines with IL-17 led to the downregulation of miR-23b, the expression of miR-23b and IL-17 in tissue resident cells seems to be mutually antagonistic12. IL-17 is mainly produced by activated T-cells, has the ability to activate other T-cells, and is also capable of triggering the inflammatory process in many other tissues/cells16,17. Involvement of miR-23b in the pathogenesis of myositic muscle was recently reported12 and the etiopathogenetic role of IL-17 in IIM has also been previously described18,19. The aim of our research was to analyze the changes in expression of miR-23b/miR-23b* in PBMCs from patients with myositis and to evaluate the importance of the IL-17/miR-23b signaling network in the PMBCs of these patients. Our results clearly show that the expression profile of miR-23b (as well as miR-23b*) is not changed in the PBMCs of IIM patients. Therefore, the regulative role of miR-23b in the development of myositis is not taking place in PMBCs. We cannot exclude involvement of this system in the local inflammatory process within the inflamed myositis muscle tissue.
Author contributions
JV and PN conceived and designed the study. MR, MS and TS carried out the research. MS prepared the first draft of the manuscript. MR and TS contributed to the experimental design and preparation of the manuscript. PN provided direct supervision of the research. All authors were involved in the revision of the draft manuscript and have agreed to the final content.
Competing interests
No competing interests were disclosed.
Grant information
This work was supported by the Internal Grant Agency of the Ministry of Health in the Czech Republic (Nr. NT 12452) and a grant from the Charles University Grant agency Nr. 621812.
Acknowledgements
We are grateful to Jitka Smekalova and Marketa Kubalkova for excellent technical assistance.
Faculty Opinions recommendedReferences
- 1.
Ceribelli A, Yao B, Dominguez-Gutierrez PR, et al.:
MicroRNAs in systemic rheumatic diseases.
Arthritis Res Ther.
2011; 13(4): 229. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 2.
Galeazzi M, Balistreri E, Giannitti C, et al.:
MicroRNAs in autoimmune rheumatic diseases.
Reumatismo.
2012; 64(1): 7–17. PubMed Abstract
| Publisher Full Text
- 3.
Basu A, Jiang X, Negrini M, et al.:
MicroRNA-mediated regulation of pancreatic cancer cell proliferation.
Oncol Lett.
2010; 1(3): 565–568. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 4.
Feng S, Cong S, Zhang X, et al.:
MicroRNA-192 targeting retinoblastoma 1 inhibits cell proliferation and induces cell apoptosis in lung cancer cells.
Nucleic Acids Res.
2011; 39(15): 6669–78. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 5.
Le MT, Teh C, Shyh-Chang N, et al.:
MicroRNA-125b is a novel negative regulator of p53.
Genes Dev.
2009; 23(7): 862–76. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 6.
Sato F, Tsuchiya S, Meltzer SJ, et al.:
MicroRNAs and epigenetics.
FEBS J.
2011; 278(10): 1598–609. PubMed Abstract
| Publisher Full Text
- 7.
O’Connell RM, Rao DS, Baltimore D:
microRNA regulation of inflammatory responses.
Annu Rev Immunol.
2012; 30: 295–312. PubMed Abstract
| Publisher Full Text
- 8.
Baltimore D, Boldin MP, O’Connell RM, et al.:
MicroRNAs: new regulators of immune cell development and function.
Nat Immunol.
2008; 9(8): 839–845. PubMed Abstract
| Publisher Full Text
- 9.
O’Connell RM, Rao DS, Chaudhuri AA, et al.:
Physiological and pathological roles for microRNAs in the immune system.
Nat Rev Immunol.
2010; 10(2): 111–122. PubMed Abstract
| Publisher Full Text
- 10.
Dai R, Ahmed SA:
MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases.
Transl Res.
2011; 157(4): 163–179. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 11.
Alevizos I, Illei GG:
MicroRNAs as biomarkers in rheumatic diseases.
Nat Rev Rheumatol.
2010; 6(7): 391–398. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 12.
Zhu S, Pan W, Song X, et al.:
The microRNA miR-23b suppresses IL-17–associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α.
Nat Med.
2012; 18(7): 1077–86. PubMed Abstract
| Publisher Full Text
- 13.
Bohan A, Peter JB:
Polymyositis and dermatomyositis (first of two parts).
N Engl J Med.
1975; 292(7): 344–7. PubMed Abstract
| Publisher Full Text
- 14.
Bohan A, Peter JB:
Polymyositis and dermatomyositis (second of two parts).
N Engl J Med.
1975; 292(8): 403–7. PubMed Abstract
| Publisher Full Text
- 15.
Chomczynski P, Sacchi N:
Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction.
Anal Biochem.
1987; 162(1): 156–159. PubMed Abstract
| Publisher Full Text
- 16.
Yao Z, Fanslow WC, Seldin MF, et al.:
Herpesvirus Saimiri Encodes a New Cytokine , IL-17 , which Binds to a Novel Cytokine Receptor.
Immunity.
1995; 3(6): 811–821. PubMed Abstract
| Publisher Full Text
- 17.
Yao Z, Painter SL, Fanslow WC, et al.:
Human il-17: A novel cytokine derived from T cells.
J Immunol.
1995; 155(12): 5483–6. PubMed Abstract
- 18.
Zhu W, Streicher K, Shen N, et al.:45
Genomic signatures characterize leukocyte infiltration in myositis muscles.
BMC medical genomics.
2012; 5: 53. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 19.
Tournadre A, Miossec P:
Interleukin-17 in inflammatory myopathies.
Curr Rheumatol Rep.
2012; 14(3): 252–6. PubMed Abstract
| Publisher Full Text

1) Report the ... Continue reading This is potentially a very interesting paper if miR-23b is selective for certain autoimmune diseases over others. There are several things that could greatly improve the data however.
1) Report the data as relative fold change using the delta delta Ct (ddCt) method. First, normalize data to endogenous control (dCt) followed by an additional normalization to the highest dCt value to obtain the ddCt. This can be easily converted to Relative fold change. Relative fold change is considered better than dCt because one can compare across platforms.
2) It is unclear why the author uses 4 genes to normalize the expression (dCt). It is more common to select 1 gene. This needs to be explained and a graph showing the Ct values of each gene should be provided.
3) For Figure 1, a follow up figure should be provided to demonstrate any possible differences that may exist amoung PM and DM patients and Healthy controls. Diseases should always be segregated.
4) For Figure 1, another follow up figure should show males and females separately to demonstrate that sex has no effect.
5) For Figure 1, the controls almost appear to have two populations- a high and low. Why is this and could some of the healthy controls possibly not be very healthy?
6) The t-test is fine; however, a Mann-Whitney test would be better for human data.
7) It may be interesting to plot miR-23b vs miR-23b* for both healthy controls and patients to see if how they relate to each other (slopes) is different between patient and healthy controls.
8) A table displaying the clinical data of the patients is something that is commonly included and would be useful to any physician.
With more data analysis and reporting of relative fold change, this paper could have some interesting results. Segregating patients into different groups and comparing to the healthy controls could greatly improve the value of the data.
1) Report the data as relative fold change using the delta delta Ct (ddCt) method. First, normalize data to endogenous control (dCt) followed by an additional normalization to the highest dCt value to obtain the ddCt. This can be easily converted to Relative fold change. Relative fold change is considered better than dCt because one can compare across platforms.
2) It is unclear why the author uses 4 genes to normalize the expression (dCt). It is more common to select 1 gene. This needs to be explained and a graph showing the Ct values of each gene should be provided.
3) For Figure 1, a follow up figure should be provided to demonstrate any possible differences that may exist amoung PM and DM patients and Healthy controls. Diseases should always be segregated.
4) For Figure 1, another follow up figure should show males and females separately to demonstrate that sex has no effect.
5) For Figure 1, the controls almost appear to have two populations- a high and low. Why is this and could some of the healthy controls possibly not be very healthy?
6) The t-test is fine; however, a Mann-Whitney test would be better for human data.
7) It may be interesting to plot miR-23b vs miR-23b* for both healthy controls and patients to see if how they relate to each other (slopes) is different between patient and healthy controls.
8) A table displaying the clinical data of the patients is something that is commonly included and would be useful to any physician.
With more data analysis and reporting of relative fold change, this paper could have some interesting results. Segregating patients into different groups and comparing to the healthy controls could greatly improve the value of the data.