Introduction
Molecular events such as mutation, deletion, recombination and gene transfer play paramount roles in shaping the evolution of prokaryotes (reviewed by1,2). As a consequence the genomes are more prone for nucleotide base compositional fluctuations. Particularly, the evolutionary forces pose a major impact on the guanine-cytosine (GC) content of bacteria at the level of genes and genomes3. Amongst all the important evolutionary forces, the impact of recombination (homologous, non-homologous or illegitimate) on the evolution of bacteria has been evidenced as the major driving force or factor of microevolution4–8. However, the rate of recombination may differ greatly amongst different bacterial species; while some species recombine more frequently to have multiple recombination events than mutations that render them weakly clonal, whereas in other species it appears to be a rare incident leading to distinct clonal lineages5,6,8,9. Studies of genetic diversity in the bacterial kingdom have shown that bacteria form clusters of genetically related strains and that extensive recombination among related clusters have been regarded as normal rather than exceptional events10. However, not every single gene is involved in recombination or horizontal gene transfer11,12.
The lineages of the genes were broadly classified into informational and operational or metabolic housekeeping genes. The informational genes include genes of translation (T), transcription (S), and replication (R) and also the ATPases, GTPases (G) and tRNA synthetases whereas the operational genes are those involved in cell operations such as amino acid synthesis (A), biosynthesis of co-factors (B), cell envelope proteins (C), energy metabolism (E), intermediary metabolism (I), fatty acid and phospholipid biosynthesis (L), nucleotide biosynthesis (N), and regulatory genes (Z)12. The operational genes are the most modular genes in the cells that are inclined to be horizontally transferred or recombined most often11,13. As a result of this behaviour of the housekeeping genes it is therefore prudent to speculate that housekeeping genes may exhibit notable molecular differences compared to the informational genes.
Campylobacter jejuni (C. jejuni), is a zoonotic pathogen that colonises the gut of a wide variety of birds and mammals and has been attributed to the majority of bacterial gastroenteritis cases in developed countries14. Most predominantly the disease is caused by C. jejuni and often the disease is self limiting, however on rare occasions there can be serious sequelae such as Guillain-Barré syndrome and reactive arthritis15. The natural competency and the plasticity of C. jejuni were not investigated until Dingle et al. (2001)16 designed an MLST scheme for C. jejuni which has subsequently been exploited to structure and investigate the association of C. jejuni populations with different hosts and the environment from which human clinical infection originated17–21. Further, Wilson et al.22 used the population genetics-phylogenetics approach to demonstrate the massive evolutionary potential of C. jejuni inferring that recombination plays a major role in the generation of diversity at twice the rate of mutation per se.
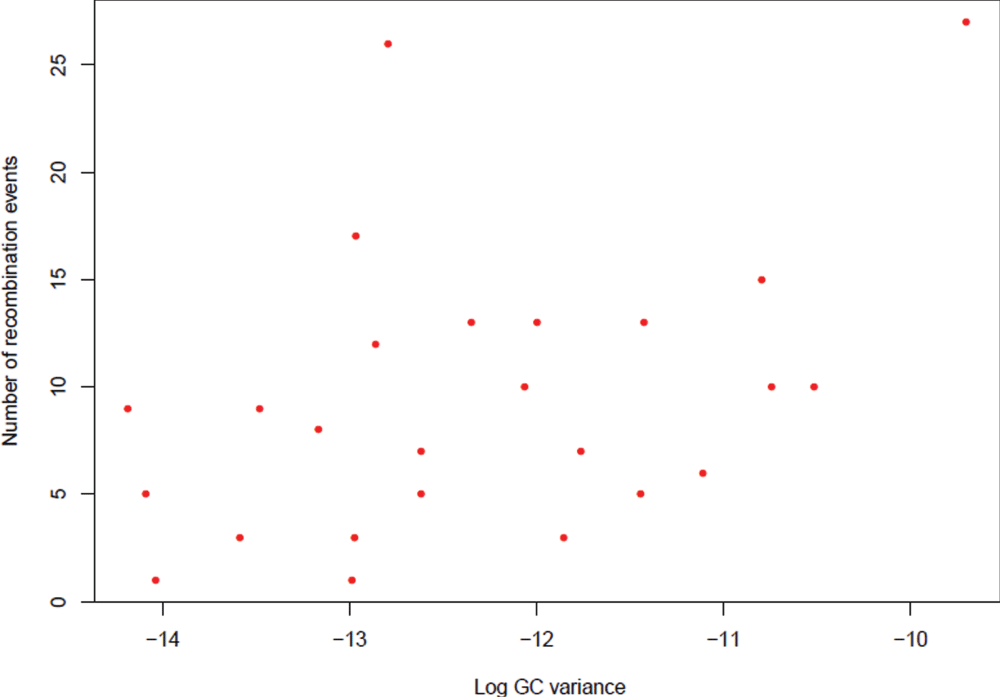
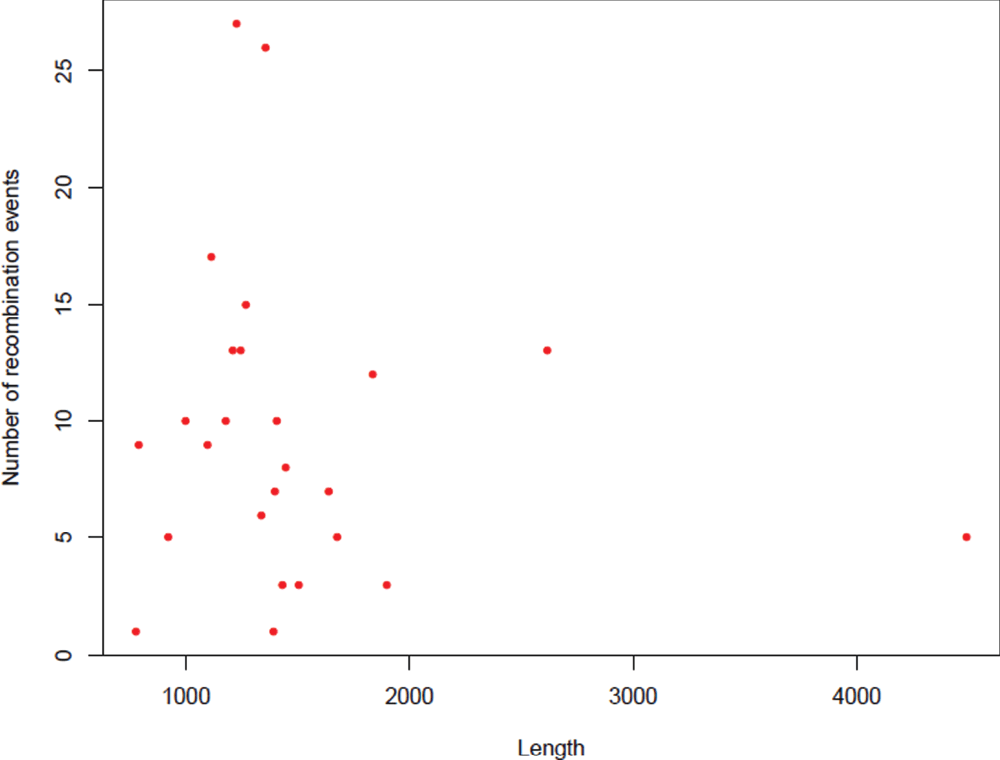
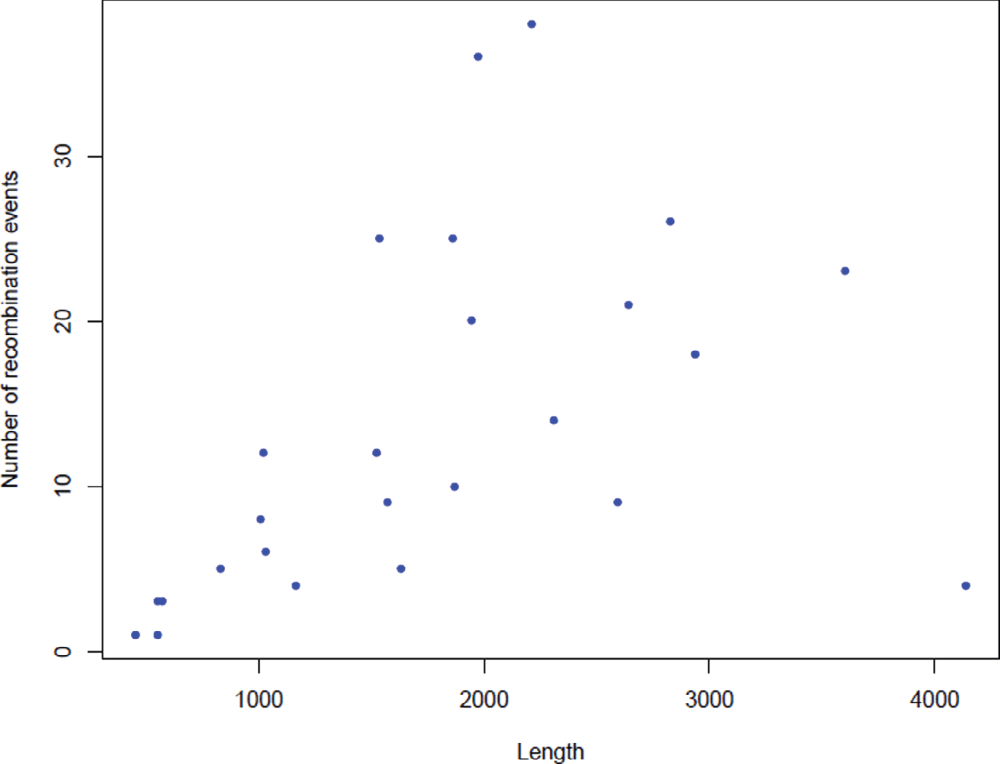
As C. jejuni is an actively recombining bacterial species, an attempt to investigate the GC variations in a subset of operational genes and informational genes was made in an effort to understand the difference between these two lineages of genes within C. jejuni genomes. The assumptions to conduct this analysis were: (1) amelioration or coalescence of GC content or the nucleotide base composition takes relatively long time23 which means that an event of recombination in a given group of genes with lack of time for coalescence will exhibit notable GC variation compared to other genes in the population; (2) housekeeping genes are relatively more prone for recombination where interspecies recombination has been demonstrated between donor and recipient DNA molecules that differ by up to 25–30% of their nucleotide sites24–26. Given the fact that C. jejuni is a competent bacterial species where generation of host adapted variants has been documented using MLST datasets by several studies27–31, it may be hypothesised that the housekeeping genes may exhibit notable differences in their GC content24; (3) Informational genes require highly stringent homology for an event of recombination to occur and recombination takes place as a part of DNA repair32 and hence these genes may not exhibit such notable GC variation even in the presence of recombination events. In order to test these hypotheses, nineteen C. jejuni genomes were analysed in this study.

Comments on this article Comments (0)