Introduction
The cell surface of most Gram-positive bacteria contains wall teichoic acid and lipoteichoic acid with a poly-alditol phosphate backbone. The remaining hydroxyls of the alditol moiety are ubiquitously modified by D-alanyl esterification or glycosylation1–3. A dlt operon, which typically codes for DltA, DltB, DltC and DltD proteins, is required for the D-alanylation of lipoteichoic acids4. D-alanylation brings in positively charged amino-groups and partially neutralizes the net negative charges of the phosphate groups on the lipoteichoic acid backbone. The reduction of D-alanyl content on the cell surface has been found to be associated with increased autolysis3,5 and susceptibility to host defense peptides and other antibiotics6,7. Impaired D-alanylation of lipoteichoic acid also reduces the ability of bacteria to colonize any surface8 and form antibiotics-resistant biofilms9. Therefore, the dlt-encoded proteins required for the lipoteichoic acid D-alanylation pathway could serve as novel targets for fighting emerging infectious diseases caused by Gram-positive pathogens10. The DltA inhibitor 5’-O-[N-(D-alanyl)-sulfamoyl]adenosine for example, designed by analogy to D-alanyl adenylate (D-Ala-AMP), has been shown to significantly suppress the growth of B. subtilis when used in combination with vancomycin11.
The D-alanyl carrier protein ligase DltA (formed by ~500 amino acid residues)4 closely resembles (~30% sequence identity) the adenylation domains (also called adenosine monophosphate (AMP)-forming domains) found in bacterial non-ribosomal peptide synthetases12. Its remote homologs include the acyl- and aryl-(coenzyme A) CoA synthetases and firefly luciferases13. As shown in Figure 1, DltA catalyzes the ATP-driven adenylation of D-alanine and the subsequent transfer of the activated D-alanyl group to the thiol group of 4’-phosphopantetheine, which is covalently bound to a serine side chain of D-alanyl carrier protein DltC (~80 amino acid residues)4. As previously shown for such one-protein-two-enzymes homologs14,15, crystal structures of two DltA proteins have also been observed in adenylation and thiolation conformations, respectively16,17 (Figure 1). The thiolation conformation of B. subtilis DltA (BsDltA)18 resembles previously determined structures of a bacterial acetate-CoA synthetase (ACS) and 4-chlorobenzoate-CoA ligase crystallized in their respective thiolation state15,19. On the other hand, the adenylation conformation of B. cereus DltA (BcDltA)17 resembles those observed in the crystal structures of the firefly luciferase PheA (phenylalanine-activating domain of the first module of the Bacillus brevis gramicidin S synthetase I), DhbE (2,3-dihydroxybenzoate activation domain from B. subtilis), a yeast acetyl-CoA synthetase and 4-chlorobenzoate-CoA ligase13,14,20–22. The two conformations captured in the crystal structures of the two closely related DltA proteins (56% sequence identity) are related by a ~146º rotation around a hinge residue Asp-399 in BcDltA (Asp-398 in BsDltA), which is a invariable residue equivalent to Asp-517 in Salmonella typhimurium acetyl-CoA synthetase23. The region surrounding this hinge residue (residues Arg-397 to Glu-413 of BcDltA green in Figure 1B) contains an important triphosphate-interacting residue Arg-39724 and a β-hairpin which serves as part of the pantetheine channel as observed in the bacterial acetyl-CoA synthetase23.
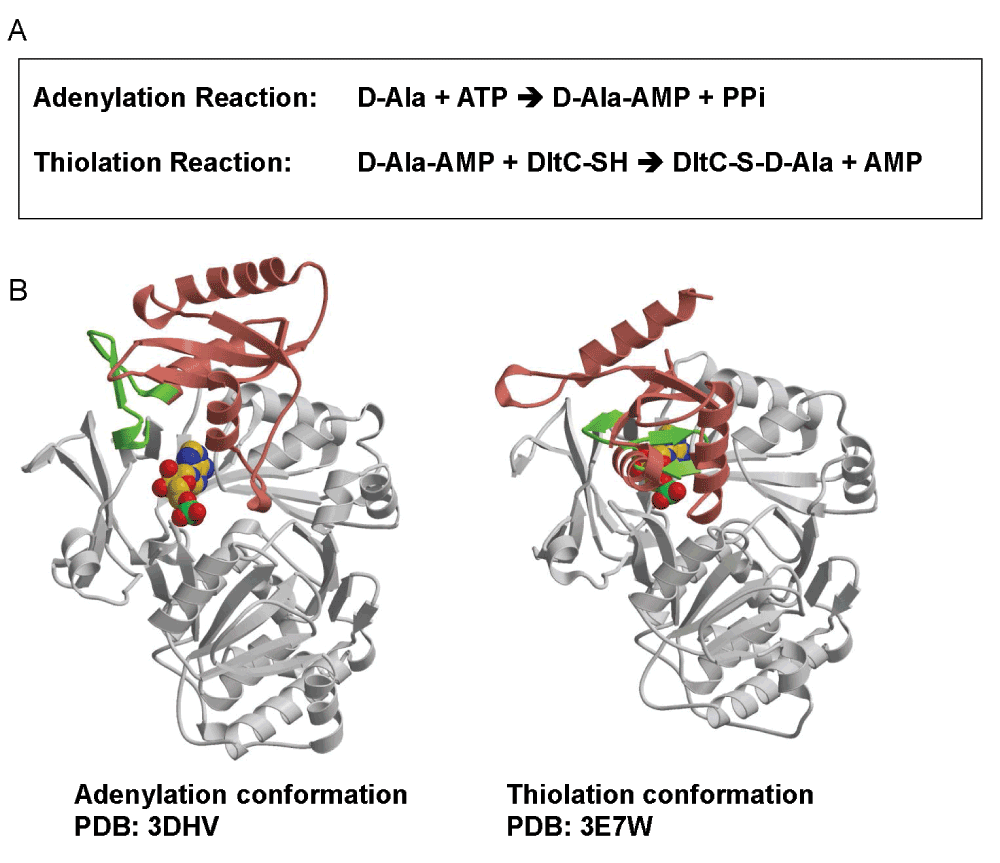
Figure 1. Structure and function of DltA.
A. A two-step reaction catalyzed by DltA. B. The adenylation and thiolation conformations of DltA. In the first adenylation reaction, D-alanine is converted into DltA-bound D-alanine adenylate (D-Ala-AMP). This reaction is catalyzed by the adenylation conformation of DltA shown as DltA/AMP complex. In the subsequent thiolation reaction, the activated alanyl group in the intermediate is transferred to the thiol group of DltC-linked 4’-phosphopantetheine (DltC-SH). The second reaction is catalyzed by the thiolation conformation of DltA shown also as DltA/AMP complex. Most part of the major N-terminal domain of DltA (residues 2 - 399) is shown in gray. The majority of the minor C-terminal domain of DltA (residues 400–504) is shown in salmon. Residues between Arg-397 and Glu-413 are highlighted in green.
We previously studied the adenylation reaction of BcDltA in the absence of any D-alanyl carrier to enable the thiolation reaction17. The resulting specificity constants (kcat/KM) for D- and L-alanine differed by merely ~3-fold, which could not explain the fact that teichoic acid is overwhelmingly modified by D- but not L-alanine25. As suggested by a study on a spectrum of DltA homologs which have residual aminoacyl-CoA synthetase activity42, CoA has been confirmed to be a substitute for the D-alanyl carrier protein DltC as the thiolation substrate of BcDltA24. Here we report further biochemical analysis of BcDltA. Noticeable differences were observed in Michaelis constants KM and turnover rates kcat. The presence of CoA, hence the enabled adenylation and thiolation cycle, enhanced the enzyme’s apparent affinities to its cognitive substrate ATP and D-alanine by approximately an order of magnitude. Since BcDltA is a slow enzyme with turnover rate less than 1 s-1, much slower than bacterial Acyl-CoA synthetases (~102 s-1)23 and 4-chlorobenzoate-CoA ligase (~10 s-1)26, the observed Michaelis constants should closely approximate the corresponding dissociation constants, and therefore provide some insight into the stability of short-lived enzyme-substrate complexes. We also determined the structure of BcDltA in the absence of any substrate. This structure is noticeably more disordered than previously reported DltA structures16,17,24, which may explain the enzyme’s lower affinity to ATP in the absence of the other two substrates. Interestingly, CoA-enhanced affinities to ATP and D-alanine imply that the thiolation substrate CoA-bound BcDltA has higher affinity to both adenylation substrates as compared to CoA-free BcDltA.
Methods
Cloning, protein preparation and crystallization
All reagents were from VWR unless specified otherwise. The wild-type as well as the C269A mutant of DltA from B. cereus was cloned for over-expression of BcDltA as described previously17. The pET28-BcDltA construct carries an Ala-1 mutation at the N-terminus and eight extra residues at the C-terminus (LEHHHHHH). The soluble fraction of BcDltA was purified by nickel-affinity chromatography followed by gel filtration. BcDltA was concentrated to ~20 mg/mL by ultra-filtration.
Crystallization and structure determination
The concentrated BcDltA protein was crystallized using the hanging drop crystallization method at a room temperature of 21°C. The optimal well solution for crystallization contained 0.1 M MgCl2, 0.5 M KCl, 16% polyethylene glycol (PEG) 3,350 (Sigma-Aldrich) and 0.05 M Hepes-NaOH buffer at pH 7.2. Each drop was composed of 1 μL of protein and 1 μL of well solution. The plate-shaped crystals grew to a maximal size of 0.4 mm × 0.3 mm × 0.05 mm in three days. Crystals were gradually transferred to stabilizing solutions composed of the crystallization well solution supplemented with 8%, 16% and 24% glycerol, soaked for 1 minute, then flash-cooled to - 173.15°C in a nitrogen stream generated by an Oxford CryoSystems device. A total of 400 0.4-degree oscillation images were acquired and processed using a Brukers Proteum-R system as already described27. The previously solved BcDltA model (PDB code 3DHV)17 was used as the starting model for two-domain rigid body refinement followed by positional refinement using Crystallography & NMR System (CNS)28. This resulting model was subjected to ten cycles of rebuilding and refinement using Arp/Warp29. The rebuilt model was iteratively rebuilt using XtalView30 and then refined using CNS28. The final model had 90.8% of the residues in the most favored regions on a Ramachandran plot. Val-301 with clear electron density and Asp-336 with blurry electron density were the only two residues found in the disfavored region. Statistics of the diffraction data, refinement and geometry are listed in Table 1. The molecular figures were generated using Molscript31 and rendered using Raster3D32. The coordinates and structure factors have been deposited in the Protein Data Bank33–35 (entry code 4PZP).
Table 1. X-ray crystallographic data collection and structure refinement statistics.
| Substrate-free BcDltA |
|---|
|
Data collection
| |
| Space group | P21
|
| Cell dimensions | |
| a, b, c (Å) | 52.9, 81.9, 59.3 |
| α, β, γ (°) | 90.0, 108.3, 90.0 |
| Resolution (Å) | 33.1–1.9 (2.00–1.90)a
|
|
bRsym
| 0.077 (0.321) |
| I/σI | 7.5 (1.8) |
| No. reflections | 32733 (1982) |
| Completeness (%) | 86.3 (37.7) |
| Redundancy | 4.3 (3.2) |
|
Refinement
| |
| Resolution (Å) | 30.0–1.9 |
| No. reflections | 32658 |
|
cRwork/Rfree
| 0.218/0.261 |
| No. atoms: | |
| Protein | 3643 |
| Water | 279 |
| Average B-factors: | |
| Protein | 22.9 |
| Water | 27.7 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.47 |
Tryptophan fluorescence measurement
The intrinsic tryptophan fluorescence of 1.0 ml 0.4 uM BcDltA solution with 0 to 2 mM ATP was acquired at a room temperature of 21°C using a PerkinElmer LS-55 fluorescence spectrometer. The excitation wavelength was 305 nm and a fluorescent emission in the 310 nm to 390 nm range was recorded. The relative fluorescence increase at 345 nm was used to quantify the ATP-bound fraction of BcDltA. Assuming that the fluorescence gain is proportional to [ATP]/(KD + [ATP]), Prism software (GraphPad Software) was used to derive the dissociation constant of ATP.
Pyrophosphate quantification assay
As previously described, pyrophosphate released from the adenylation reaction is broken down into phosphate by pyrophosphatase17. The resulting phosphate was quantified by a dye solution containing 0.033% w/v Malachite Green, 1.3% w/v ammonium molybdate and 1.0 M HCl36. The 200 μL reaction solutions contained 5 μM BcDltA, 0.1 M KCl, 0.01 M MgCl2, 0.05 M Tris-Hepes buffer at pH 7.2, 5 unit/mL of inorganic pyrophosphatase from baker’s yeast (Sigma-Aldrich) and specified concentrations of D-alanine, ATP and CoA (Sigma-Aldrich). A volume of 25 μL of reaction solution was retrieved every 3 or 5 minutes and mixed thoroughly with 475 μL of the dye solution. The absorption at a wavelength of 620 nm was recorded after 90 seconds. The initial rates (1/2 of the phosphate concentration increase per minute) of the adenylation reaction were derived from the time courses of phosphate accumulation. The correlation between initial reaction rate and substrate concentration was fitted with Michaelis-Menten equation using the Prism software (GraphPad Software).
Thiol quantification assay
The free thiol group of CoA was quantified as described previously24 by a dye solution composed of 1 mM 5,5’-dithio-bis (2-nitrobenzoic acid) (DTBN), or Ellman’s reagent (Sigma-Aldrich) and 50 mM Tris-EDTA solution at pH 8.0. Absorption at a wavelength of 412 nm was used to quantify the concentration of free thiol group. The reaction rate was derived by the rate of thiol depletion. The correlation between initial reaction rate and substrate concentration was also fitted with Michaelis-Menten equation using the Prism software.
Results
Crystal structure of substrate-free BcDltA
The same DltA protein from B. cereus with a C-terminal hexahistidyl fusion tag used in our previous crystallographic studies on BcDltA17,24 was crystallized in the absence of ATP, D-alanine or CoA. One crystal diffracted to 1.9 Å resolution and belonged to space group P21 (Table 1), the same space group as in previously reported crystals of BcDltA in complex with D-alanine adenylate17 and with ATP24. Despite having 8 Å shorter crystallographic a axis, 5 Å shorter b axis and 5° smaller β angle than the previously reported crystal of DltA/D-alanine adenylate complex, the structure was successfully solved by rigid-body refinement using the previously determined BcDltA structure17 (PDB code 3DHV) as the starting model. The 504-residue BcDltA structure can be divided into two domains (Figure 2): an N-terminal major domain from the N-terminus to Asp-399, and a C-terminal minor domain from residue 400 to the C-terminus. The disposition of the two domains in the substrate-free BcDltA structure remains similar to that of the starting model (Figure 2A). The electron density map indicated several disordered regions (Ser-153 to Pro-159, Pro-363 to Glu-367, Arg-397 to Glu-413, Lys-433 to Tyr-440) with the corresponding regions in the starting model highlighted in magenta in Figure 2B. The first disordered region is part of a highly conserved P-loop (Thr-152 to Lys-160) found in homologous AMP-forming proteins37–39. Due to its similar amino acid composition (glycine, serine, threonine and lysine) to that of P-loop or Walker A motif found in ATPases and GTPases40, this loop has long been thought to catalyze the adenylation reaction. In the crystal structure of human medium-chain acyl-CoA synthetase in complex with ATP, this loop intimately interacts with the β- and γ-phosphates of the ATP substrate41. Functional relevance of regions 363–367 and 433–440 are unknown. The former is located at a 2-turn helix at the surface of the N-terminal domain (Figure 2B). The latter is interacting with the longest disordered inter-domain region in this structure (397–413) which contains several key elements of DltA. Arg-397 has been observed to interact with the β-phosphate of ATP and to play an important role in catalysis24. Asp-399, equivalent to Asp-398 in BsDltA, serves as the hinge residue for domain rotation. As observed for the equivalent Asp-402 of 4-Chlorobenzoate-CoA ligase15, the rotation around main-chain single bonds in this hinge residue could account for a 146° swing of the C-terminal domain as we compared the crystallized adenylation conformation of BcDltA and thiolation domain of BsDltA (Figure 1, bottom with the disordered inter-domain linker in green). The equivalent main-chain atoms in the N-terminal domains of BcDltA in adenylation conformation (PDB entry 3DHV) and BsDltA in thiolation conformation (PDB entry 3E7W) are superposed with a root-mean-deviation of 0.97 Å, and the deviation for the C-terminal domain was 1.00 Å. These values indicate that there is no dramatic conformational change within each domain in addition to the 146° rotation around the hinge aspartate residue. The C-terminal part of this flexible inter-domain region also contains a β-hairpin which has been observed to interact with CoA in homologous acetyl-CoA synthetase23.
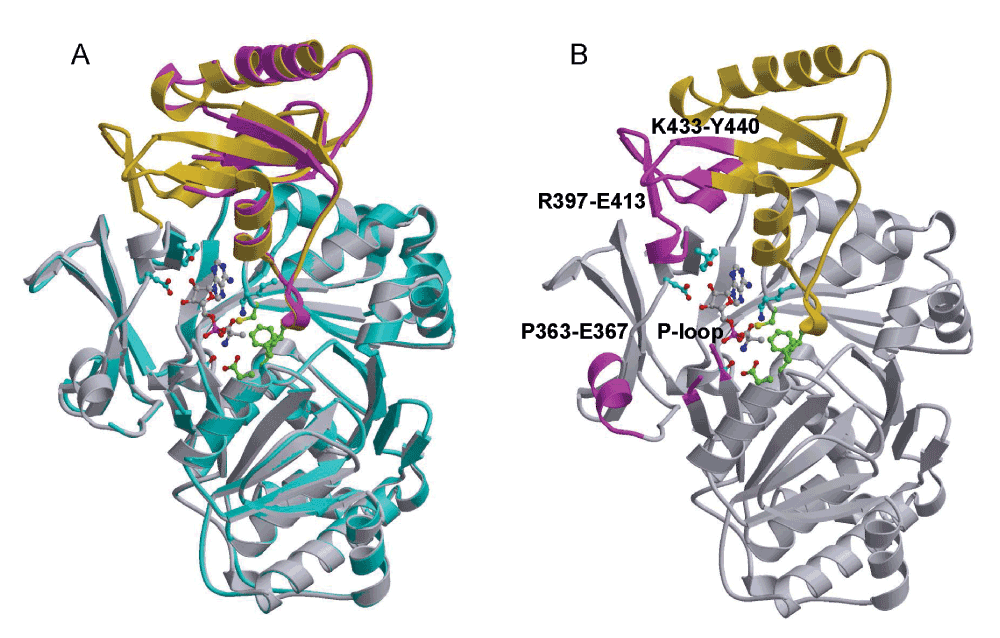
Figure 2. Structure of DltA in the absence of substrate.
The ribbons representation of previously reported BcDltA structure (PDB entry: 3DHV) is shown with D-alanine-adenylate and surrounding side-chains in ball-and-stick model. The N-terminal domain is shown in gray, and the C-terminal domain in gold. A. The substrate-free structure of BcDltA is superimposed on the BcDltA/D-Ala-AMP complex. The N- and C-terminal domains are colored in cyan and magenta, respectively. B. The four corresponding regions of the BcDltA/D-Ala-AMP complex, which are disordered in the substrate-free form of BcDltA, are highlighted in magenta.
KM and kcat of BcDltA in the presence and absence of D-alanyl carrier CoA
In our previous study, we have verified that CoA can mimic D-alanyl carrier protein DltC24, as also discovered for DltA homologs42. In that study, we have observed that the reaction rate is increased by nearly an order of magnitude by the presence of saturating concentration of CoA, which is explained by the faster release of the thiolation product rather than by release of the adenylation intermediate. In order to get a comprehensive understanding of the effects by CoA as the DltC mimic, we further studied the enzymatic properties of BcDltA in the presence of a saturating concentration (5 mM) of ATP or D-alanine, and in the absence of CoA or in the presence of a saturating concentration (5 mM) of CoA. The reaction rates derived from the pyrophosphate accumulation assay and the thiol depletion assay were similar (Figure 4). The thiolation assay was noticeably noisier than the pyrophosphate assay and we therefore limit the discussion to KM and kcat values derived from the pyrophosphate assay. Somewhat unexpectedly, BcDltA showed much higher apparent affinity, or decreased KM value, towards ATP (0.46 mM to 0.01 mM) and D-alanine (1.1 mM to 0.03 mM) in the presence of 5 mM CoA (Figure 3 and Figure 4, Table 2). On the contrary, the apparent affinity towards L-alanine decreased in the presence of CoA, with KM increased from 14.4 mM to 109 mM (Figure 4 and Table 2).
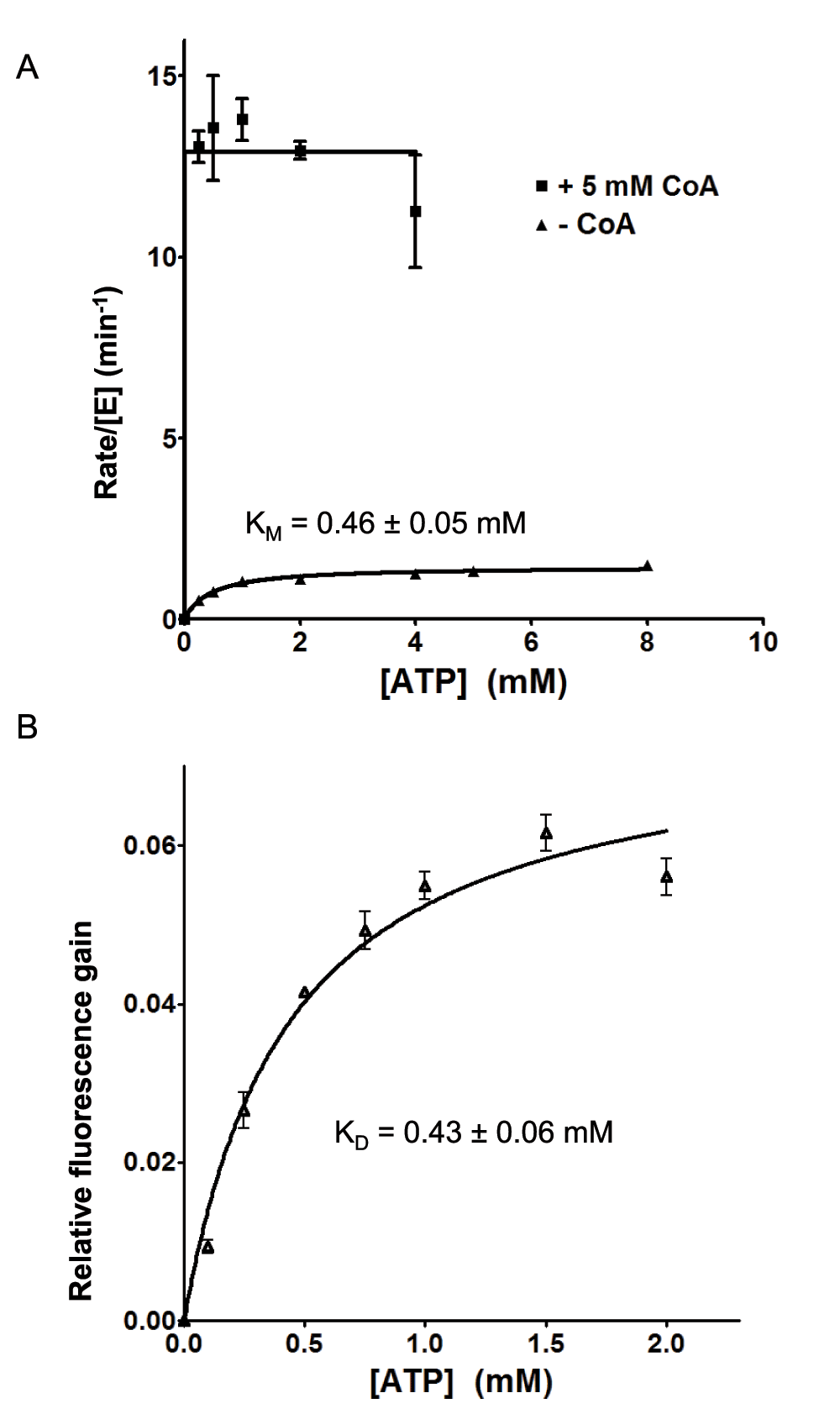
Figure 3. Rate of pyrophosphate release and ATP-induced fluorescence gain.
The reaction solutions contained 0.005 mM wild-type BcDltA, specified concentrations of ATP and CoA, 5 mM D-alanine, 0.1 M KCl, 0.01 M MgCl2, 0.05 M Hepes-NaOH buffer at pH 7.2, 5 unit/mL of yeast inorganic pyrophosphatase. A. The initial rates of pyrophosphate accumulation divided by the BcDltA concentration are shown. The reaction rates in the presence of CoA are taken from a previous study24. B. Relative fluorescence gains as a fraction of the fluorescence intensity in the absence of ATP are shown.
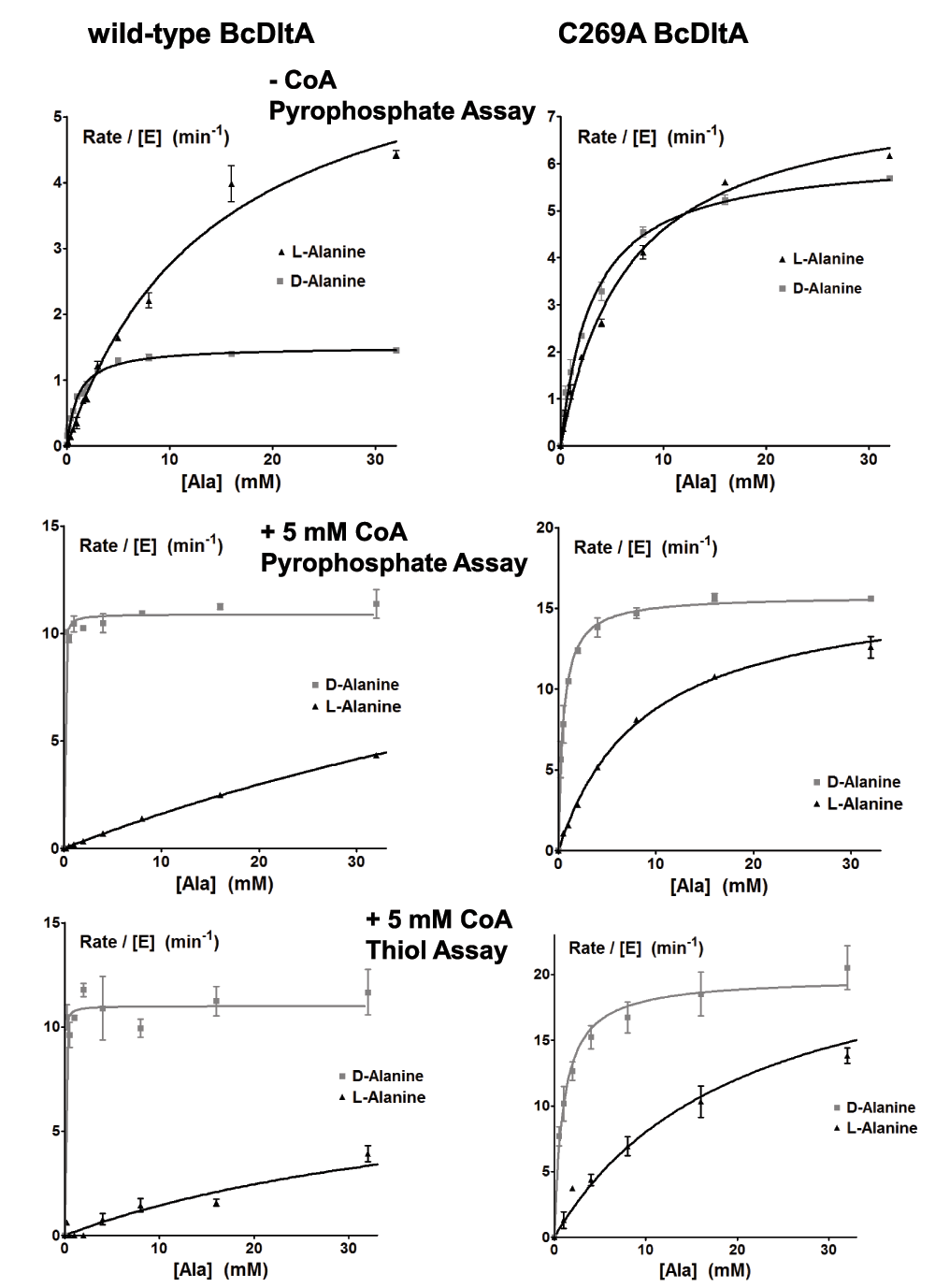
Figure 4. Rate of pyrophosphate release and thiol depletion.
The reaction solutions contained 0.005 mM wild-type BcDltA, 5 mM D-alanine, 0.1 M KCl, 0.01 M MgCl2, 0.05 M Hepes-NaOH buffer at pH 7.2, 5 unit/mL of yeast inorganic pyrophosphatase and specified concentrations of ATP, alanine and CoA. Reaction rates for wild-type BcDltA are shown on the left, and those for the C269A mutant protein are shown on the right. The reaction rates in the absence of CoA are taken from a previous study17.
Table 2. Kinetic data for DltA protein from B. cereus.
| BcDltA (substrate) | KM (× 10-3 M) | kcat (min-1) | kcat/KM (× 103 M-1·min-1) |
|---|
|
Pyrophosphate assay (5 mM ATP, - CoA) (previous work17)
|
| Wild-type (D-Ala) | 1.1 ± 0.2 | 1.5 ± 0.1 | 1.4 ± 0.2 |
| Wild-type (L-Ala) | 14.4 ± 1.6 | 6.7 ± 0.4 | 0.47 ± 0.05 |
| C269A (D-Ala) | 3.1 ± 0.3 | 6.2 ± 0.2 | 2.0 ± 0.2 |
| C269A (L-Ala) | 6.6 ± 0.5 | 7.6 ± 0.3 | 1.2 ± 0.1 |
|
Pyrophosphate assay (5 mM ATP, 5 mM CoA) (this work)
|
| Wild-type (D-Ala) | 0.03 ± 0.01 | 10.9 ± 0.2 | 363 ± 100 |
| Wild-type (L-Ala) | 109 ± 7 | 19.3 ± 0.7 | 0.18 ± 0.02 |
| C269A (D-Ala) | 0.50 ± 0.05 | 15.8 ± 0.3 | 32 ± 3 |
| C269A (L-Ala) | 8.8 ± 0.5 | 16.5 ± 0.4 | 1.9 ± 0.2 |
|
Thiol assay (5 mM ATP, 5 mM CoA) (this work)
|
| Wild-type (D-Ala) | 0.02 ± 0.02 | 11.0 ± 0.3 | 550 ± 200 |
| Wild-type (L-Ala) | 48 ± 12 | 8.4 ± 1.0 | 0.18 ± 0.02 |
| C269A (D-Ala) | 1.0 ± 0.2 | 19.8 ± 0.8 | 19.8 ± 0.9 |
| C269A (L-Ala) | 20 ± 3 | 24.3 ± 1.6 | 1.2 ± 0.2 |
|
Pyrophosphate assay (5 mM D-Ala, 5 mM CoA) (previous work24)
|
| Wild-type (ATP) | 0.01 ± 0.02 | 12.9 ± 0.6 | 1290 ± 2000 |
|
Pyrophosphate assay (5 mM D-Ala, - CoA) (this work)
|
| Wild-type (ATP) | 0.46 ± 0.05 | 1.46 ± 0.04 | 3.2 ± 0.04 |
Relaxed D-alanine preference by the C269A BcDltA mutant protein
The side-chain of Cys-269 sits at the bottom of the D-alanine-binding pocket which may make VDW clash with the methyl side-chain of L-Alanine16,17. We also studied the effect of CoA on D- and L-alanine preference of the C269A mutant of BcDltA (Figure 4 and Table 2). In the presence and absence of CoA, the C269A protein showed relaxed preference for D-alanine over L-alanine. As observed for the wild-type protein, CoA also enhances the D-alanine preference of this BcDltA mutant protein. However, the CoA-induced changes in Michaelis constant KM were less dramatic (3.1 mM to 0.50 mM for D-alanine, 6.6 mM to 8.8 mM for L-alanine) than those observed for the wild-type protein.
ATP binding by BcDltA
Change in tryptophan fluorescence of BcDltA was minimal in the presence of ATP in the 10 micromolar range. We then found that at an excitation wavelength of 305 nm, the absorption of up to 2 mM of ATP was negligible and there was fluorescence gain associated with increasing concentration of ATP. The fluorescence gain-derived dissociation constant KD for ATP (0.43 mM) was similar to the Michaelis constant KM (0.46 mM) derived in the absence of CoA (Figure 3). There was no detectable fluorescence change to derive KD values for D-alanine. For CoA, we were not able to isolate fluorescence change from absorption by CoA in millimolar concentration.
Discussion
DltA strongly prefers D- over L-alanine
Bacteria selectively incorporate D- over L-alanine in cell wall components. There appears to be no exception in ubiquitous esterification of lipoteichoic acids by alanine25. The enantiomer selectivity of BcDltA observed in the absence of any D-alanyl carrier17, however, has been intriguingly mediocre. The newly acquired kinetic data in the presence of saturating CoA, a DltC mimic, shows that the kcat values are less than 2-fold different for D- and L-alanine (10.9 and 19.3 min-1) while the KM values are more than 1000-fold different favouring D-alanine over L-alanine (0.03 and 109 mM). Adding the fact that the intracellular concentration of D-alanine (in the order of 102 μM) is approximately 10-fold more abundant than the L-enantiomer (in the order of 101 μM)43, DltA functioning at a saturating concentration of the D-alanyl carrier protein DltC would favor the ligation of the D-enantiomer by approximately 4 orders of magnitude. Such striking enantiomer selectivity is consistent with the much lower L-alanine content found in lipoteichoic acid25. A recent study has shown that the dlt operon is induced by cell envelope stress such as acidic pH and antibiotics44. It is possible that stress-induced expression of DltC may reach a saturating concentration for interacting with DltA and therefore ensure the almost exclusive enantiomer selectivity as observed in the presence of the DltC mimic. The comparison between the kinetic properties of the wild-type and the C269A mutant proteins also supports the notion that Cys-269, and its equivalence in other DltA proteins, contributes to the enantiomer selectivity of DltA16,17.
Thiolation conformation of DltA is compatible with adenylation substrates
It is satisfying to observe the stringent D-alanine preference. At the same time, it is also puzzling to appreciate that such selectivity on the chirality of alanine, a substrate of the adenylation reaction, can only be achieved in the presence of CoA, a substrate of the thiolation reaction. The intrinsic fluorescence of tryptophan in BcDltA enabled us to derive the dissociation constant KD for ATP in the absence of other substrates. The new form of crystal structure in the absence of any substrate is noticeably more disordered than the previously observed adenylation conformation of BcDltA17 and thiolation conformation of BsDltA16. The longest such disordered region is between Arg-397 and Glu-413, which contains the inter-domain hinge residue Asp-399, interacts with β-phosphate of ATP, and forms the pantetheine channel. Possibly, this important region remains disordered in the presence of saturating D-alanine, therefore providing an explanation for the similar values between the above mentioned KD (0.43 mM) and the KM (0.46 mM) for ATP, in the presence of saturating D-alanine but in absence of CoA. The more disordered nature of the substrate-free conformation of BcDltA also implies that the previously observed adenylation and thiolation conformations are intrinsically unstable unless stabilized by the interaction with one or more substrates. This structural feature of BcDltA likely explains the relatively low sub-millimolar affinity for ATP, since part of the stabilizing BcDltA-ATP interactions would be used to compensate the cost of establishing the adenylation conformation of the protein.
BcDltA is a very slow enzyme. Unless the substrate dissociation step happens to be extremely slow as well, we could approximate the observed KM values to the KD values of corresponding BcDltA-substrate intermediates, and therefore enable reasoning in the context of structural stability of such intermediates. As such, we reasoned that the approximately one order of magnitude difference in KM values for the adenylation substrates ATP and D-alanine observed in the presence and absence of the thiolation substrate may imply the existence of a quadruple intermediate of the DltA enzyme in complex with all three substrates, which may be markedly different from a ternary intermediate of BcDltA with the two adenylation substrates.
We then resorted to three-dimensional model building so as to answer the question on which of the adenylation and thiolation conformations may be compatible with binding to all three substrates. The BcDltA/D-alanine-adenylate complex (PDB entry 3DHV)17 was chosen as the adenylation conformation and as the reference set of atomic coordinates. The adenylated intermediate was dissected to generate the D-alanine model. The N-terminal domain of the BcDltA/ATP complex (PDB entry 3FCE)24, which is also in the adenylation conformation, was superposed on the reference set to orient the ATP substrate. The N-terminal domain of the BsDltA/AMP complex (PDB entry 3E7W) was also superposed on the reference set to derive the re-oriented thiolation conformation. Main-chain atoms equivalent to those interacting with AMP in BcDltA (270–272, 292–299) in the quadruple complex of acetyl-CoA synthetase in its thiolation conformation (PDB entry 2P2F)23 were superimposed on the reference set to orient the CoA model. In the adenylation conformation (Figure 5A, the pantetheine channel is apparently blocked by the main-chain atoms immediately preceding the catalytic Lys-492 of BcDltA (Lys-491 of BsDltA). Although we could not completely rule out the possibility that an allosteric site for CoA exists, no such site has ever been observed for this superfamily of enzymes. Therefore, a quadruple complex in the adenylation conformation is unlikely. The thiolation conformation (Figure 5B), on the other hand, appears to be compatible with binding ATP so long as Arg-408 of BcDltA (Arg-407 of BsDltA) adopts another rotamer. It has been previously observed that the homologous acetyl-CoA synthetase in its thiolation conformation binds AMP, acetate and CoA23, and that BsDltA in its thiolation conformation binds AMP and appears to have a well-formed D-alanine-binding pocket16. These structural evidences seem to suggest that a quadruple intermediate may form with the enzyme in its thiolation conformation but not in the adenylation conformation. Since the thiolation conformation has an AMP-binding site and its N-terminal domain, which provides most of the ATP-interacting residues such as the P-loop and Arg-397, remains essentially identical to that that in the adenylation conformation, it is not surprising that only one arginine side-chain is required to adopt another rotamer to accommodate ATP. This arginine residue (Arg-408 of BcDltA) forms a salt bridge with the divalent cation-anchoring side-chain of Glu-298 in BcDltA (Glu-297 in BsDltA)24, which appears to be an important structural feature to modulate the conformational change16. Another arginine residue (Arg-397 of BcDltA, Arg-396 of BsDltA) also seems to facilitate the conformational change. It forms a salt bridge with the hinge aspartate residue in the thiolation conformation. In the adenylation conformation, this arginine side-chain adopts a more extended rotamer and forms a salt bridge with the β-phosphate group of ATP (Figure 5A). ATP binding would result in re-orientation of both arginine residues and disruption of two salt bridges, thus mobilizing the thiolation conformation.
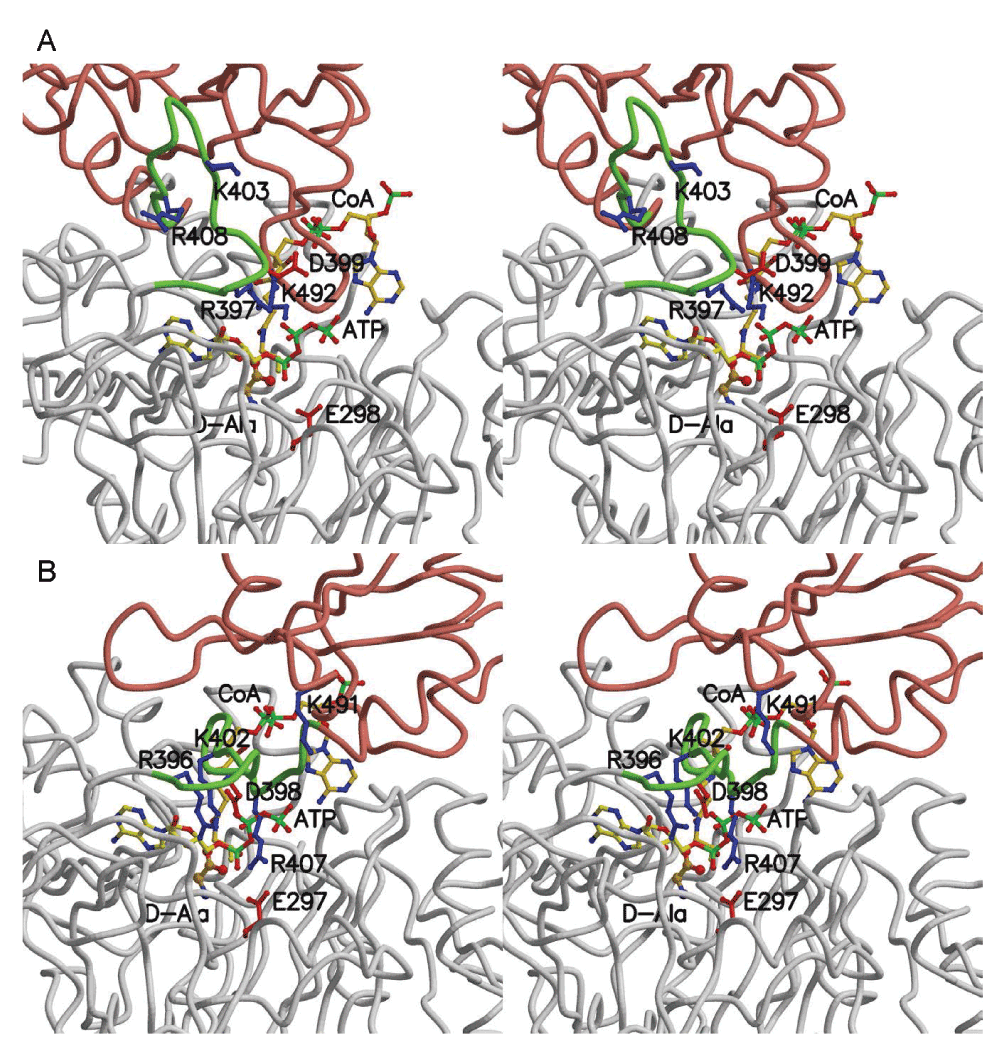
Figure 5. Structural model of DltA in complex with ATP, D-alanine and CoA.
Cα traces of the two modelled quadruple complexes are shown in stereo. Except for the green-colored region between Arg-397 and Glu-413, the N- and C-terminal domains are colored in gray and salmon, respectively. The three substrates and selected side-chains are shown in ball-and-stick model. A. The quadruple complex in adenylation conformation. Residue numbers in BcDltA are shown. B. The quadruple complex in thiolation conformation. Residue numbers in BsDltA are shown.
Structural basis for CoA-enhanced affinity for ATP
The majority of ATP-binding elements lie in the N-terminal domain which remains similar in both adenylation and thiolation conformations. The most significant structural feature in the C-terminal domain for binding ATP is the catalytic Lys-492 of BcDltA (Lys-491 of BsDltA) in the adenylation conformation. In the thiolation conformation, the catalytic residue is replaced by Lys-403 (Lys-402 of BsDltA) (Figure 5). In addition, the thiolation conformation is more ordered than the substrate-free conformation. The favourable DltA/ATP interactions may no longer be used to compensate the energetic cost of stabilizing the disordered hinge region. Therefore the affinity for ATP by the CoA-bound BcDltA in its thiolation conformation should be higher than substrate-free BcDltA.
Structural basis for CoA-enhanced enantiomer selectivity for D-alanine
For both the wild-type and C269A mutant BcDltA, the presence of saturating CoA increases the apparent affinity for D-alanine while decreasing affinity for L-alanine. The majority of alanine-interacting residues lie in the N-terminal domain. The amino group of D-alanine is stabilized by Asp-197 of BcDltA (Asp-196 of BsDltA). Cys-269 of BcDltA (Cys-268 of BsDltA) lies close to Cα of D-alanine and serves as one major determinant of enantiomer selectivity. The carboxylate group of D-alanine is stabilized by Lys-492 in the adenylation conformation or by Lys-403 in the thiolation conformation. At the end of the pantetheine channel, Phe-196 of BcDltA (Phe-195 of BsDltA) adopts different rotamers in the two conformations. In the adenylation conformation of BcDltA/D-alanine adenylate, the shortest distance from Phe-196 side-chain to methyl group of D-alanine is 4.15 Å. In the modelled quadruple complex in the thiolation conformation, the Phe-196 side-chain does not contact D-alanine but the thiol group of CoA lies in closer proximity of D-alanine side-chain (3.34 Å), as expected from the pending thiolation reaction between the thiol group of CoA and the carbonyl group of D-alanine-adenylate. On the other hand, the tighter alanine-binding pocket may exert stronger VDW repulsion toward L-alanine, thus lowering further the affinity for the wrong enantiomer. Interestingly, Cys-269 also has a thiol group. The removal of this group in the C269A mutant of BcDltA reduces the affinity for D-alanine and increases the affinity for L-alanine by approximately one order of magnitude, which is almost exactly the opposite to the effect of introducing the thiol group of CoA in the alanine-binding pocket. Since sulphur atoms are larger and more inducible than the second-period elements carbon and oxygen, it is not surprising that the VDW interaction involving a thiol group makes a significant impact on the stability of enzyme/substrate complex.
Hypothesized enzymatic cycle of DltA
Intracellular concentration of D-alanine43 generally exceeds the KM value in the presence of saturating ATP and CoA. Typical intracellular concentration of ATP lies in the millimolar range45, exceeding the KM value for ATP as well. The concentration of the possibly stress-induced DltC44 may also reach saturating level in bacteria when such stress is present. As implied by the CoA-triggered dramatic change in KM values for ATP and D-alanine, the two adenylation substrates are likely incorporated by the enzyme with pre-bound CoA rather than with merely the other adenylation substrate. In addition, three-dimensional modelling suggests the CoA-bound state of the enzyme can only exist in its thiolation conformation. We therefore hypothesize a two-conformation model for the enzymatic cycle catalyzed by DltA (Figure 6) in the presence of saturating concentration of the three substrates. Our model differs from the three-conformation model proposed for DltA and adenylation domains in non-ribosomal peptide synthetase16 which includes a third substrate-free conformation. This model also differs from the two-conformation model proposed for and for 4-chlorobenzoate-CoA ligase26,46 which includes substrate-free state of the enzyme. The two previously proposed models are consistent with a typical Ping-Pong mechanism. Neither a substrate-free conformation nor a substrate-free state of the enzyme is required in our model for the enzymatic cycle of DltA once the protein enters the reaction cycle. The adenylation reaction starts from a ternary complex with ATP and D-alanine and proceeds with the release of pyrophosphate. A domain rotation around the aspartate hinge residue follows transforming into the thiolation conformation which binds CoA, or DltC in bacteria, and catalyzes the thiolation reaction with D-alanine by displacing AMP. In our model, the resulting complex with the two thiolation products proceeds with AMP release, D-alanine-CoA/CoA exchange, D-alanine binding, and ATP binding. In the quadruple complex, the ATP-bound magnesium ion would replace Arg-408 of BcDltA in forming a bridge with Glu-298, as observed in the crystal structure BcDltA/ATP complex24. The disruption of the Arg-408 to Glu-298 salt bridge would destabilize the thiolation conformation and facilitate a reverse domain rotation and release of CoA which is not compatible with the adenylation conformation. The hypothesized D-alanine-CoA/CoA exchange step is the central piece of this enzymatic cycle, which reflects the finding that CoA affects DltA’s apparent affinities with D-alanine and ATP. If DltA were to become substrate-free for a long enough period of time, it would form a ternary complex with ATP and D-alanine, which contradicts the observed effect by the thiolation substrate CoA. It is worth noting that the sequence of D-alanine and ATP binding is uncertain. Similarly, AMP release has to occur before ATP binding but not necessarily before exchange with CoA or D-alanine binding.
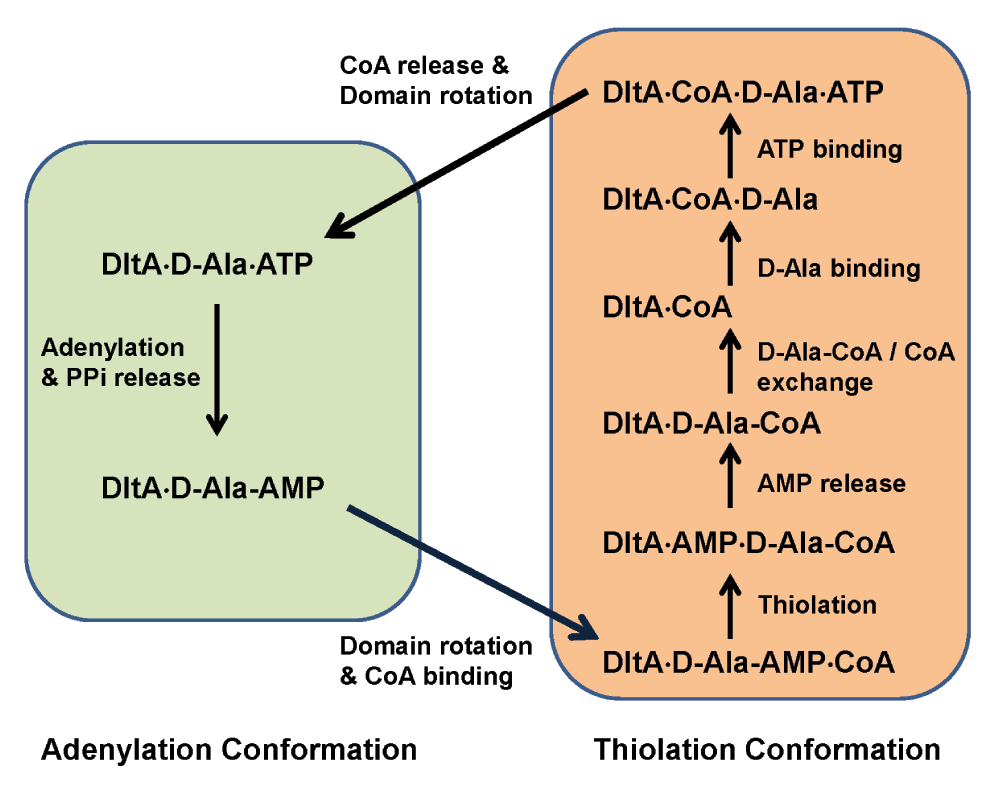
Figure 6. Hypothetical enzymatic cycle of DltA.
Non-covalent DltA complexes are shown in text. Forward reaction, substrate binding and release steps are shown by arrows. Steps fulfilled by the adenylation conformation are shown in the green box, and those by the thiolation conformation in brown. The thioester between D-Ala and CoA (D-Ala-CoA) is the final product.
The hypothesized enzymatic cycle involves a second CoA binding step, and a CoA release step in addition to the Ping-Pong mechanism previously proposed for DltA and its homologs. Both additional steps seem unnecessary for the enzymatic reaction itself, but are required to explain our enzymatic data and are consistent with the three-dimensional models of BcDltA and BsDltA. A Ping-Pong mechanism would require that the binding of CoA and the binding of either adenylation substrate be uncompetitive, and therefore the apparent KM and kcat values for either adenylation substrate both would become larger at higher CoA concentration. While the kcat values for both ATP and D-alanine did become larger at saturating CoA concentration, the KM values actually became smaller, therefore contradicting the typical Ping-Pong mechanism. Another property of aminoacyl-CoA synthetases including DltA is the inhibitory effect of CoA at high concentration42, which is difficult to explain by a typical Ping-Pong mechanism unless the enzymatic cycle includes an additional CoA-dissociation step as in our model. The closest homologs of DltA are amino acid-activation domains found in non-ribosomal peptide synthetases42. Similar to DltA, these homologs also pass the adenylated intermediate to the 4’-phosphopantetheine group attached to a serine residue on a peptide carrier domain. It is possible that such amino acid activation domains may also act as DltA.
The extra binding step for 4’-phosphopantetheine D-alanyl carrier could serve as the sensor for the availability of the carrier. The maximum rate catalyzed by BcDltA is approximately seven times faster in the presence of CoA than in its absence, with respective kcat values 10.9 min-1 and 1.5 min-1. Moreover, the intracellular concentration of D-alanine is typically found in the 100 micromolar range43, which lies above the KM value for D-alanine in the presence of CoA (~30 µM) but below the KM value in the absence of CoA (1100 µM) (Table 2). The reaction rate should become slower by approximately 100 fold when the thiol carrier is absent. It is worth noting that the adenylated D-alanine intermediate generated by the adenylation reaction is not covalently attached to the enzyme and could be released and wasted when the thiolation substrate is absent. The significant slowing down of the adenylation reaction in the absence of the 4’-phosphopantetheine carrier therefore provides a biological advantage.
Data availability
figshare: Data of pyrophosphate release, ATP-induced fluorescence and thiol depletion for DltA in Bacillus cereus, http://dx.doi.org/10.6084/m9.figshare.101848947
Author contributions
YL conceived the study, designed the experiments, determined the crystal structure, and wrote the manuscript. LD carried out cloning, expression, crystallization and biochemical assays, and approved the manuscript for publication.
Competing interests
No competing interests were disclosed.
Grant information
This work is supported by a Saskatchewan Health Research Foundation Phase 3 Team Grant to the Molecular Design Research Group at University of Saskatchewan, and by a Natural Sciences and Engineering Research Council Discovery Grant 261981-2010 to YL.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgements
We thank Drs. Gabriele Schatte and Wilson Quail for assistance with the X-ray facility at the Saskatchewan Structural Sciences Centre.
Faculty Opinions recommendedReferences
- 1.
Armstrong JJ, Baddiley J, Buchanan JG, et al.:
Composition of teichoic acids from a number of bacterial walls.
Nature.
1959; 184: 247–248. PubMed Abstract
| Publisher Full Text
- 2.
Hyyrylainen HL, Vitikainen M, Thwaite J, et al.:
D-Alanine substitution of teichoic acids as a modulator of protein folding and stability at the cytoplasmic membrane/cell wall interface of Bacillus subtilis.
J Biol Chem.
2000; 275(35): 26696–26703. PubMed Abstract
| Publisher Full Text
- 3.
Fischer W:
Physiology of lipoteichoic acids in bacteria.
Adv Microb Physiol.
1988; 29: 233–302. PubMed Abstract
- 4.
Perego M, Glaser P, Minutello A, et al.:
Incorporation of D-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation.
J Biol Chem.
1995; 270(26): 15598–15606. PubMed Abstract
- 5.
Wecke J, Perego M, Fischer W:
D-alanine deprivation of Bacillus subtilis teichoic acids is without effect on cell growth and morphology but affects the autolytic activity.
Microb Drug Resist.
1996; 2(1): 123–129. PubMed Abstract
| Publisher Full Text
- 6.
Peschel A, Otto M, Jack RW, et al.:
Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides.
J Biol Chem.
1999; 274(13): 8405–8410. PubMed Abstract
- 7.
Kristian SA, Lauth X, Nizet V, et al.:
Alanylation of teichoic acids protects Staphylococcus aureus against Toll-like receptor 2-dependent host defense in a mouse tissue cage infection model.
J Infect Dis.
2003; 188(3): 414–423. PubMed Abstract
| Publisher Full Text
- 8.
Gross M, Cramton SE, Gotz F, et al.:
Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces.
Infect Immun.
2001; 69(5): 3423–3426. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 9.
Gotz F:
Staphylococcus and biofilms.
Mol Microbiol.
2002; 43(6): 1367–1378. PubMed Abstract
| Publisher Full Text
- 10.
Neuhaus FC, Baddiley J:
A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in gram-positive bacteria.
Microbiol Mol Biol Rev.
2003; 67(4): 686–723. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 11.
May JJ, Finking R, Wiegeshoff F, et al.:
Inhibition of the D-alanine:D-alanyl carrier protein ligase from Bacillus subtilis increases the bacterium's susceptibility to antibiotics that target the cell wall.
FEBS J.
2005; 272(12): 2993–3003. PubMed Abstract
| Publisher Full Text
- 12.
Stachelhaus T, Mootz HD, Marahiel MA:
The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases.
Chem Biol.
1999; 6(8): 493–505. PubMed Abstract
| Publisher Full Text
- 13.
Conti E, Franks NP, Brick P:
Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes.
Structure.
1996; 4(3): 287–298. PubMed Abstract
| Publisher Full Text
- 14.
Gulick AM, Lu X, Dunaway-Mariano D:
Crystal structure of 4-chlorobenzoate:CoA ligase/synthetase in the unliganded and aryl substrate-bound states.
Biochemistry.
2004; 43(27): 8670–8679. PubMed Abstract
| Publisher Full Text
- 15.
Reger AS, Wu R, Dunaway-Mariano D, et al.:
Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
Biochemistry.
2008; 47(31): 8016–8025. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 16.
Yonus H, Neumann P, Zimmermann S, et al.:
Crystal structure of DltA: implications for the reaction mechanism of non-ribosomal peptide synthetase (NRPS) adenylation domains.
J Biol Chem.
2008; 283(47): 32484–91. PubMed Abstract
| Publisher Full Text
- 17.
Du L, He Y, Luo Y:
Crystal structure and enantiomer selection by D-alanyl carrier protein ligase DltA from Bacillus cereus.
Biochemistry.
2008; 47(44): 11473–11480. PubMed Abstract
| Publisher Full Text
- 18.
Yonus H, Neumann P, Zimmermann S, et al.:
Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains.
J Biol Chem.
2008; 283(47): 32484–32491. PubMed Abstract
| Publisher Full Text
- 19.
Gulick AM, Starai VJ, Horswill AR, et al.:
The 1.75 A crystal structure of acetyl-CoA synthetase bound to adenosine-5'-propylphosphate and coenzyme A.
Biochemistry.
2003; 42(10): 2866–2873. PubMed Abstract
| Publisher Full Text
- 20.
Conti E, Stachelhaus T, Marahiel MA, et al.:
Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S.
EMBO J.
1997; 16(14): 4174–4183. PubMed Abstract
| Free Full Text
- 21.
May JJ, Kessler N, Marahiel MA, et al.:
Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc Natl Acad Sci U S A.
2002; 99(19): 12120–12125. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 22.
Jogl G, Tong L:
Crystal structure of yeast acetyl-coenzyme A synthetase in complex with AMP.
Biochemistry.
2004; 43(6): 1425–1431. PubMed Abstract
| Publisher Full Text
- 23.
Reger AS, Carney JM, Gulick AM:
Biochemical and crystallographic analysis of substrate binding and conformational changes in acetyl-CoA synthetase.
Biochemistry.
2007; 46(22): 6536–6546. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 24.
Osman KT, Du L, He Y, et al.:
Crystal structure of Bacillus cereus D-alanyl carrier protein ligase (DltA) in complex with ATP.
J Mol Biol.
2009; 388(2): 345–355. PubMed Abstract
| Publisher Full Text
- 25.
Neuhaus FC, Linzer R, Reusch VM Jr:
Biosynthesis of membrane teichoic acid: role of the D-alanine-activating enzyme and D-alanine: membrane acceptor ligase.
Ann N Y Acad Sci.
1974; 235(0): 502–518. PubMed Abstract
| Publisher Full Text
- 26.
Wu R, Cao J, Lu X, et al.:
Mechanism of 4-chlorobenzoate:coenzyme a ligase catalysis.
Biochemistry.
2008; 47(31): 8026–8039. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 27.
Wu Y, He Y, Moya IA, et al.:
Crystal structure of archaeal recombinase RadA: a snapshot of its extended conformation.
Mol Cell.
2004; 15(3): 423–435. PubMed Abstract
| Publisher Full Text
- 28.
Brunger AT, Adams PD, Clore GM, et al.:
Crystallography & NMR system: A new software suite for macromolecular structure determination.
Acta Crystallogr D Biol Crystallogr.
1998; 54(Pt 5): 905–921. PubMed Abstract
| Publisher Full Text
- 29.
Perrakis A, Morris R, Lamzin VS:
Automated protein model building combined with iterative structure refinement.
Nat Struct Biol.
1999; 6(5): 458–463. PubMed Abstract
| Publisher Full Text
- 30.
McRee DE:
XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density.
J Struct Biol.
1999; 125(2–3): 156–165. PubMed Abstract
| Publisher Full Text
- 31.
Kraulis P:
MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures.
J Appl Cryst.
1991; 24: 946–950. Publisher Full Text
- 32.
Bacon DJ, Anderson WF:
A Fast Algorithm for Rendering Space-Filling Molecule Pictures'.
J Mol Graph.
1988; 6(4): 219–220. Publisher Full Text
- 33.
Berman HM, Westbrook J, Feng Z, et al.:
The Protein Data Bank.
Nucleic Acids Res.
2000; 28(1): 235–242. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 34.
Berman H, Henrick K, Nakamura H:
Announcing the worldwide Protein Data Bank.
Nat Struct Biol.
2003; 10(12): 980. PubMed Abstract
| Publisher Full Text
- 35.
Bernstein FC, Koetzle TF, Williams GJ, et al.:
The Protein Data Bank. A computer-based archival file for macromolecular structures.
Eur J Biochem.
1977; 80(2): 319–324. PubMed Abstract
| Publisher Full Text
- 36.
Itaya K, Ui M:
A new micromethod for the colorimetric determination of inorganic phosphate.
Clin Chim Acta.
1966; 14(3): 361–366. PubMed Abstract
| Publisher Full Text
- 37.
Chang KH, Xiang H, Dunaway-Mariano D:
Acyl-adenylate motif of the acyl-adenylate/thioester-forming enzyme superfamily: a site-directed mutagenesis study with the Pseudomonas sp. strain CBS3 4-chlorobenzoate:coenzyme A ligase.
Biochemistry.
1997; 36(50): 15650–15659. PubMed Abstract
| Publisher Full Text
- 38.
Stuible H, Buttner D, Ehlting J, et al.:
Mutational analysis of 4-coumarate:CoA ligase identifies functionally important amino acids and verifies its close relationship to other adenylate-forming enzymes.
FEBS Lett.
2000; 467(1): 117–122. PubMed Abstract
| Publisher Full Text
- 39.
Horswill AR, Escalante-Semerena JC:
Characterization of the propionyl-CoA synthetase (PrpE) enzyme of Salmonella enterica: residue Lys592 is required for propionyl-AMP synthesis.
Biochemistry.
2002; 41(7): 2379–2387. PubMed Abstract
| Publisher Full Text
- 40.
Walker JE, Saraste M, Runswick MJ, et al.:
Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold.
EMBO J.
1982; 1(8): 945–951. PubMed Abstract
| Free Full Text
- 41.
Kochan G, Pilka ES, von Delft F, et al.:
Structural snapshots for the conformation-dependent catalysis by human medium-chain acyl-coenzyme A synthetase ACSM2A.
J Mol Biol.
2009; 388(5): 997–1008. PubMed Abstract
| Publisher Full Text
- 42.
Linne U, Schafer A, Stubbs MT, et al.:
Aminoacyl-coenzyme A synthesis catalyzed by adenylation domains.
FEBS Lett.
2007; 581(5): 905–910. PubMed Abstract
| Publisher Full Text
- 43.
Manning JM, Merrifield NE, Jones WM, et al.:
Inhibition of bacterial growth by beta-chloro-D-alanine.
Proc Natl Acad Sci U S A.
1974; 71(2): 417–421. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 44.
McCormick NE, Halperin SA, Lee SF:
Regulation of D-alanylation of lipoteichoic acid in Streptococcus gordonii.
Microbiology.
2011; 157(Pt 8): 2248–2256. PubMed Abstract
| Publisher Full Text
- 45.
Beis I, Newsholme EA:
The contents of adenine nucleotides, phosphagens and some glycolytic intermediates in resting muscles from vertebrates and invertebrates.
Biochem J.
1975; 152(1): 23–32. PubMed Abstract
| Free Full Text
- 46.
Wu R, Reger AS, Lu X, et al.:
The mechanism of domain alternation in the acyl-adenylate forming ligase superfamily member 4-chlorobenzoate: coenzyme A ligase.
Biochemistry.
2009; 48(19): 4115–4125. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 47.
Liqin Du, Yu Luo:
Data of pyrophosphate release, ATP-induced fluorescence and thiol depletion for DltA in Bacillus cereus.
Figshare.
2014. Data Source






Comments on this article Comments (0)