Keywords
Neisseria meningitidis; meningococcus; meningitis; serogroup W; factor H binding protein; NadA, NHBA
Neisseria meningitidis; meningococcus; meningitis; serogroup W; factor H binding protein; NadA, NHBA
Neisseria meningitidis is a major cause of bacterial meningitis in the African Meningitis Belt1. Between 1993 and 2012, nearly 1 million suspected cases were reported with 100,000 deaths, and 80% of the cases were caused by serogroup A2. Following the introduction of the serogroup A polysaccharide conjugate vaccine MenAfriVac™ in 2010, the incidence of group A disease decreased, but outbreaks of meningitis due to other meningococcal serogroups, in particular serogroup W, continue to occur1,3. Serogroup W was responsible for an epidemic of around 13,000 cases of meningitis in Burkina Faso in 20024 and contributed to a total of 639 deaths in 2012 in the same country5. Around 40% of infected people who develop sepsis die and survivors often suffer from limb loss, cognitive dysfunction, brain damage or visual impairment.
An approach towards developing a broadly-protective meningococcal vaccine for Africa is based on the use of subcapsular antigens included in GMMA (Generalized Modules for Membrane Antigens). GMMA are outer membrane blebs from bacteria genetically engineered to release large quantities of membrane vesicles, which are enriched in outer membrane proteins. Other strain modifications are included to increase safety and immunogenicity by the up-regulation of immunogenic antigens6,7. GMMA from genetically-engineered strains with up-regulated expression of meningococcal factor H binding protein (fHbp) have been shown to provide broad protection against African meningococcal isolates from different serogroups7. Other outer membrane antigens that have been shown to induce the production of bactericidal antibodies include PorA, Neisserial adhesin A (NadA)8 and Neisserial heparin-binding antigen (NHBA)9.
To help determine the potential coverage of these antigens in a GMMA-based vaccine for Africa, we investigated their genetic diversity in serogroup W carriage and disease isolates from Burkina Faso and Ghana collected between 2003 and 2009. These two countries have suffered repeatedly from meningococcal meningitis outbreaks4,10. Focusing on isolates collected over a period of years from a defined geographic region provides the opportunity to monitor the dynamics, variation and diversity of surface-exposed antigens over time.
The N. meningitidis isolates investigated in this study were collected in the Kassena-Nankana District (KND) of Ghana and in the Nouna Health District (NHD) in the Kossi region of Burkina Faso. Case strains were isolated from the cerebrospinal fluid of meningitis patients, and carriage strains were isolated from throat swabs collected in the context of longitudinal carriage surveys. Isolation and characterization of the strains has previously been described10–13. Ethical clearance was obtained from the Ethics Committee of the War Memorial Hospital/Navrongo Health Research Centre in Ghana and the Ministry of Health and Local Ethics Committee of the Centre de Recherche en Santé de Nouna in Burkina Faso. Informed consent was obtained from all study participants. The 31 N. meningitidis carriage (n=21) and disease isolates (n=10) used in this study are described in the Table 1. The isolates were collected from Burkina Faso (n=8) and Ghana (n=23) during the period 2003–2009. The isolates were stored frozen in 10% skimmed milk at -80°C until analysis. The isolates were molecularly characterized with respect to fHbp, porA variable regions (VR), nadA and nhbA genes by sequencing. A subset of these isolates was also analysed for their fHbp expression level.
Molecular characterization was performed on these isolates by PCR amplification and sequencing of fHbp, porA, nadA and nhbA.
The selected strains were sub-cultured on GC agar plates (Becton Dickinson, Franklin Lakes, NJ, USA) and incubated overnight at 37°C, 5% CO2. A loop-full of cells was resuspended in 500 μL sterile water and boiled for 10 minutes. The samples were pelleted at 17,900 g for 5 minutes in a microcentrifuge (Eppendorf). Genomic DNA was purified using an Invitrogen PureLink Genomic DNA kit (Invitrogen, San Diego, California, USA) according to the manufacturer’s instructions. The genes encoding fHbp, PorA VRs, NadA and NHBA were PCR amplified from all isolates using the primers described in Table 2. The final PCR reaction contained: 0.5 mM deoxynucleotide triphosphates, 5 U/mL Taq DNA polymerase, 1× Thermopol Reaction buffer (all New England BioLabs, Ipswich, USA), 1 µM primer solution (Sigma-Aldrich, St. Louis, Missouri, USA) and 100 ng of genomic DNA quantified with a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, USA). The PCR was performed using the Applied Biosystems GeneAmp PCR System 9700 (Applied Biosystems, Foster City, USA) with maximum ramping speeds using conditions described in Table 3. PCR products were separated by gel electrophoresis using a 0.8% Tris base, acetic acid and EDTA (TAE) agarose gel (BioRad Laboratories, Hercules, USA). PCR products were purified using the PureLink PCR Purification Kit (Invitrogen) according to the manufacturer’s instructions. The DNA amount was measured using the NanoDrop ND-1000 Spectrophotometer.
| Target gene | Primer designation | 5′-3′ nucleotide sequence | Reference |
|---|---|---|---|
| fHbp | A1 (Fw) B2 (Rv) | GACCTGCCTCATTGAT CGGTAAATTATCGTGTTCGTACGGC | [17] [17] |
| porA | 210 (Fw) H (Rv) EI (Fw) 103L (Rv) | ATGCGAAAAAAACTTACCGCCCTC CGCATATTTAAAGGCATAG CCAGCCAGGCCATTGATCC AACGGATACGTCTTGCTC | [27] This study This study [27] |
| nadA | NadAF (Fw) NadAR (Rv) | AACACTTTCCATCCAAAG TTACCACTCGTAATTGACG | [23] [23] |
| nhbA | Fw Rv | GGCGTTCAGACGGCATATTTTTACA GGTTTATCAACTGATGCGGACTTGA | [20] [20] |
| fHbp | porA | nadA | nhbA | |
|---|---|---|---|---|
PCR profile | 94ºC, 4 minutes 35 cycles: 94ºC, 40 seconds 58ºC, 40 seconds 68ºC, 40 seconds Final extension: 72°C, 5 minutes | 94ºC, 5 minutes 30 cycles: 94ºC, 1 minute 55ºC, 1 minute 72ºC, 30 seconds Final extension: 72°C, 5 minutes | 94ºC, 5 minutes 30 cycles: 94ºC, 1 minute 55ºC, 1 minute 72ºC, 1 minute 30 seconds Final extension: 72°C, 5 minutes | 94ºC, 4 minutes 35 cycles: 94ºC, 1 minute 55ºC, 1 minute 72ºC, 1 minute Final extension: 72°C, 5 minutes |
| Reference | [17] | [27] | [23] | [20] |
The primers used for porA VR1 sequencing were 210 and 103L (Table 2). We designed primers EI and H for sequencing of the VR2 region, by aligning the conserved regions upstream and downstream of VR2 using the alignment program Clustal W (http://www.ebi.ac.uk/Tools/msa/clustalw2/). PorA sequences from the following strains of different serogroups were used for the alignment: MC58 (GenBank accession number AE002098.2), Z2491 (AL157959.1), 053442 (CP000381.1), FAM18 (AM421808.1), M6190 (AEQF01000026.1), M13399 (AEQG01000023.1) and alpha 14 (AM889136.1) using Uniprot. The sequences were read at the Novartis Vaccines-Cellular Microbiology and Bioinformatics Unit Automated DNA Sequencing Facility, Siena, Italy, on an ABI 3730 DNA Analyzer. Sequences were analyzed using the Simmonics program (version 1.6) and Chromas (version 2.01). fHbp ID, porA VR and nhbA alleles were identified using the online Neisseria Sequence Typing database (http://pubmlst.org/neisseria). nadA sample and reference sequences were exported into the MEGA software package (version 5)14 and aligned for the construction of phylogenetic trees using the maximum likelihood method with the general time-reversible model of evolution and correction for partial deletion of gaps (GenBank accession numbers: nadA1 FJ619641.1; nadA2 GQ302859.1; nadA3 JN166979.1; nadA4 FJ619644.1; nadA5 FJ619645.1). All trees were un-rooted.
For 17 isolates labelled with an asterisk in Table 1, Western blot analysis of the fHbp expression level in whole cell samples was performed as described by Seib et al.15. The strains were sub-cultured on GC agar and incubated overnight at 37°C with 5% CO2. A 7 mL aliquot of Mueller-Hinton broth (Becton Dickinson, Franklin Lakes, NJ, USA) supplemented with 0.25% glucose (Sigma-Aldrich) was inoculated with single colonies to an optical density at 600 nm (OD600) of 0.12–0.16. The suspensions were incubated at 37°C with 5% CO2 to an OD600 of 0.6 corresponding to approximately 1.8×108 cfu/ml (exponential growth phase). The cells from 1 mL of culture were collected by centrifugation at 17,900 g for 5 min in a microcentrifuge (Eppendorf), re-suspended in 100 μL phosphate buffered saline (PBS) and heat inactivated in a water bath at 56°C for 1 hour. Protein concentrations of the lysates were determined using a Lowry protein assay kit (BioRad Laboratories, Hercules, USA) with bovine serum albumin (Sigma-Aldrich) as a standard. The fHbp amounts were estimated by SDS-PAGE and Western Blot. To 100 µL heat inactivated sample we added 100 µL SDS sample buffer (Invitrogen) and 10 µL of each sample was loaded on the gel. Recombinant fHbp (rfHbp) v.2 of 500, 250, 125 and 60 ng was used as standard. Positive and negative controls were whole cell lysates from N. meningitidis group B strain 8047 expressing fHbp v.2 and the isogenic fHbp knock-out mutant. Proteins were transferred to a nitrocellulose membrane (Invitrogen) using the iBlot system (Invitrogen). After blocking overnight in 3% milk powder in PBS (Merck, Whitehouse station, NJ, USA) at 4°C, fHbp proteins were detected with 1 μg/mL anti-fHbp mouse monoclonal antibody JAR31 (IgG2b) raised against recombinant fHbp v.3 ID 28, which shows cross-reactivity against most fHbp v.2 peptides16. The secondary antibody used was 1μg/mL of a horseradish peroxidase-labelled anti-mouse IgG (Invitrogen). The membranes were developed using SuperSignal WestPico Chemiluminescent Substrate (ThermoScientific, Waltham, Massachusetts, USA) according to manufacturer’s instructions, and the signal was detected with Amersham Hyperfilm ECL (GE Healthcare, Little Chalfont, UK). The amount of fHbp expressed by each isolate compared to the standard rfHbp was determined by densitometric analysis for three biological replicates using the ImageQuant 400 gel documentation system (GE Healthcare). The expression of fHbp by the test isolates was reported as percentages of the amount of fHbp expressed by bacterial cells compared to the reference strain, known to express relatively high amounts of fHbp v.217.
PorA is an immunodominant antigen in N. meningitidis, but multiple subtypes exist with little cross-protection between meningococci expressing different PorA subtypes. fHbp can be divided into three antigenic variants, each of which is divided into sub-variants. Individual sequences are classified by a peptide ID number. Within each variant group, cross-protection is observed18. From the typing analysis of the fHbp and porA genes, all serogroup W isolates tested expressed fHbp variant 2, ID 22 (isolates from Burkina Faso) or 23 (isolates from Ghana), which differ by one amino acid, and PorA subtype P1.5,2 (Figure 1 and Supplemental File). Despite the limited number of isolates studied, these results suggest that between Burkina Faso and Ghana, which share a common border, there has been conservation of fHbp and PorA antigens among W isolates over a period of seven years.
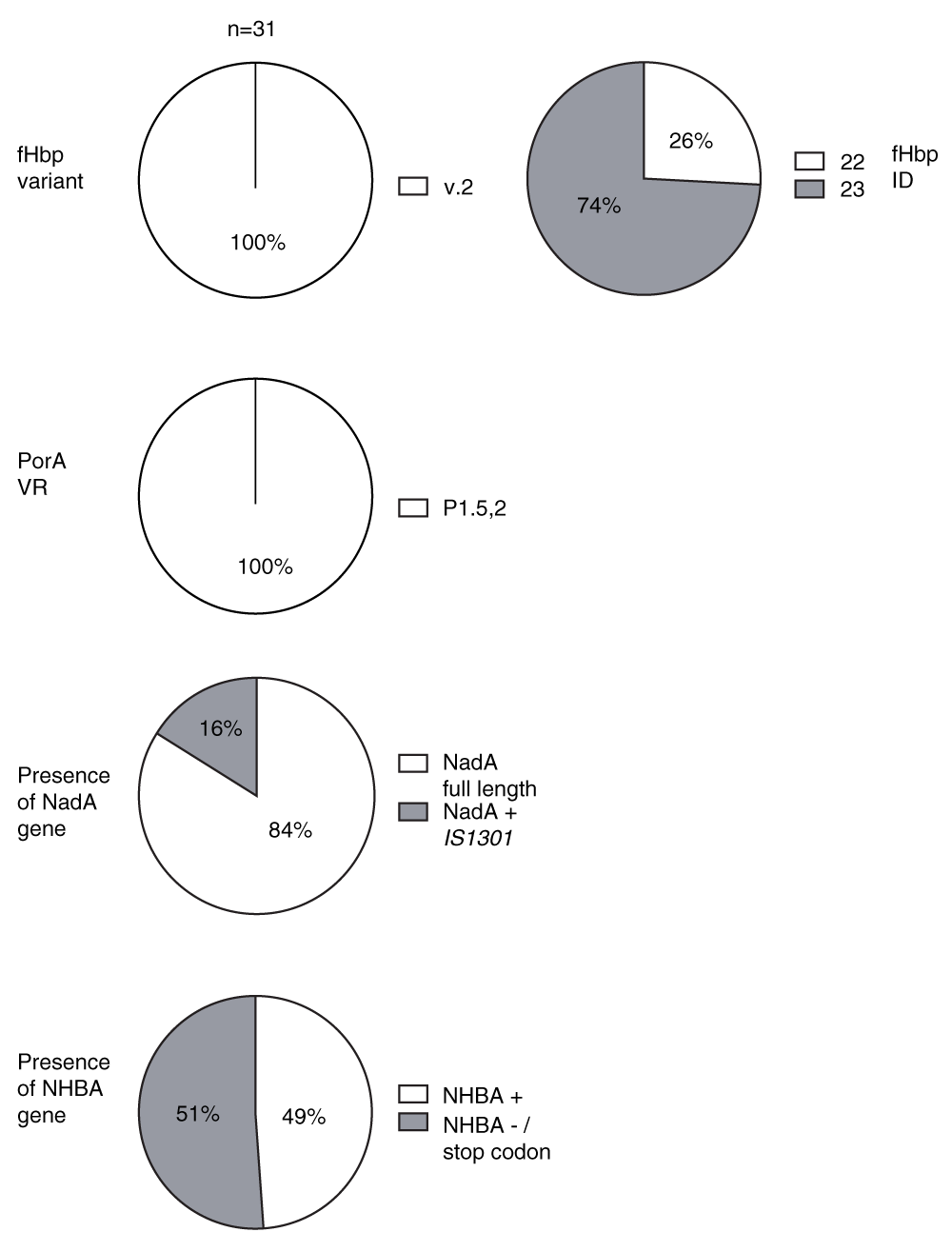
The fHbp variant group is designated according to the classification proposed by Masignani et al.17. FHbp sequence ID, PorA subtype, nadA and nhbA allele were determined by sequence query on http://pubmlst.org/neisseria. Each isolate was typed by PCR amplification of each respective gene and sequence analysis using bioinformatics software Simmonics, Mega 5 and Chromas. NadA + IS1301: Strains with NadA encoding gene containing insertion sequence IS1301. NHBA-/stop codon: Strains lacking the NHBA encoding gene or having nhbA with stop codon.
The level of fHbp protein expression can affect susceptibility of meningococci to anti-fHbp antibodies. High expressers of fHbp are generally more susceptible to killing than low expressers19. We measured fHbp expression in 17 isolates. We selected 4 out of 8 (50%) strains from Burkina Faso and 13 out 23 (56%) strains from Ghana for fHbp expression analysis. These were selected to cover isolates from different years including the oldest and newest strains. Within this group of strains selected, n=2 (50%) of the strains from Burkina Faso and n=7 (53%) of the strains from Ghana were case isolates, while the remainder were carriage isolates. We prepared whole cell extracts of the serogroup W test strains and the serogroup B reference strain and compared fHbp levels with defined amounts of a fHbp v.2 protein standard by Western blot and densitometry measurement (Dataset 1). Expression level of the reference serogroup B strain 8047 was set to 100% and levels of expression of the serogroup W strains were compared with the reference strain. Isolates with means below 33% of the reference strain were classified as low expressers while isolates with expression above 100% were categorized as high expressers. Those with mean fHbp expression between 33–100% were considered intermediate expressers. The expression of fHbp among the W isolates was variable, ranging from 50–152%, compared to the reference serogroup B strain 8047, with 41% of the isolates expressing equal or higher levels of fHbp compared to the reference strain (Figure 2). There was no significant difference in fHbp expression between case and carrier isolates studied (P=0.74, Mann Whitney U test). This indicates that levels of fHbp protein on the bacterial surface can vary among strains collected from a relatively small region and expressing the same fHbp ID.
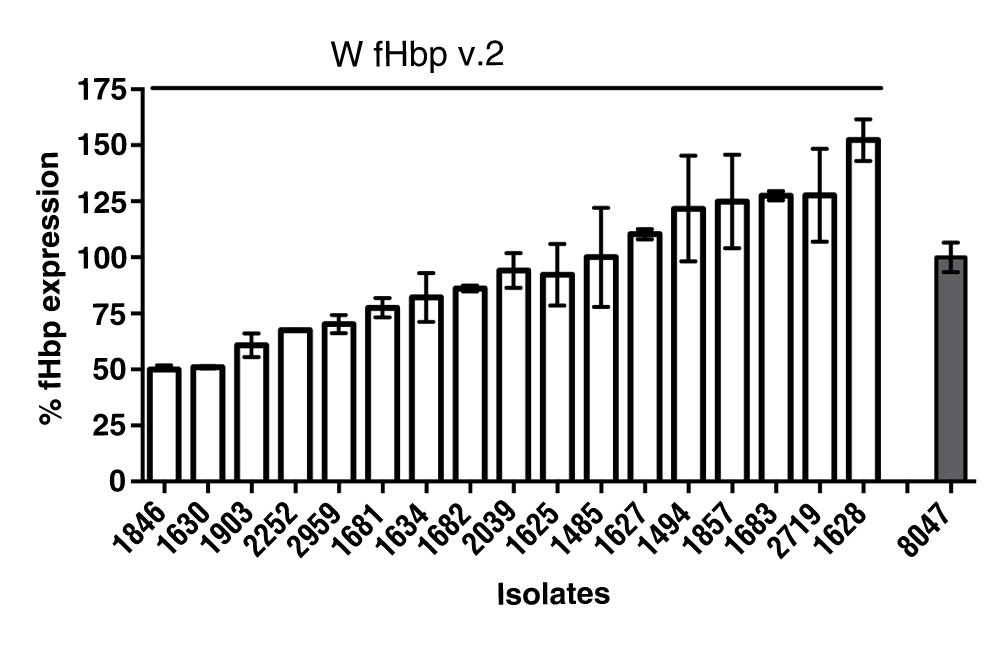
Bars represent the mean percentage from three biological replicates compared with the expression of fHbp of the reference group B strain 8047, a high expresser of fHbp variant 2 ID 77, which was set at 100%. Isolates with means below 33% were classified as low expressers while isolates with expression above 100% were categorized as high expressers. Values between 33–100% were considered as intermediate expression. Bars represent standard errors.
NadA and NHBA induce the production of bactericidal antibodies against N. meningitidis serogroup B strains. Wang et al. found that nadA was not present among a small number (n=13) of W isolates tested as part of an analysis of 896 serogroup B, C, Y and W isolates from the USA, while nhbA was present in 92% of W isolates20. Among the African W strains investigated in this study, the nadA allele 3 was present in 26/31 (84%) of isolates (Figure 1). Among the remaining five W isolates (1 case, 4 carrier isolate), PCR amplification across the nadA site gave a 2 kb product instead of the expected 1 kb product. Western blotting using whole cell lysate and polyclonal mouse anti-NadA allele 3 antibody indicated that these isolates did not express NadA (Dataset 1). Sequencing of this fragment confirmed the presence of the insertion sequence IS1301. This 842-bp mobile genetic element is known to cause a number of effects including insertions and deletions that result in silent mutations, knock-out of gene expression or regulation of downstream-located genes. For example, insertion of IS1301 into the capsular siaA gene mediates loss of encapsulation resulting in increased adherence and entry of meningococci into epithelial cells21,22. 17 out of 21 carrier (81%) and 9 out of 10 (90%) case isolates had a nadA gene. Previous reports found that nadA is present in about 50% of group B case isolates, but underrepresented in carrier isolates23.
NhbA was present in 30/31 (94%) of the meningococcal isolates studied. However, genetic sequencing in these isolates revealed a stop codon for 15/30 isolates, which has not previously been reported. The alleles of the remaining strains were identified as allele 17 (Supplemental File) using the Neisseria typing database available at http://pubmlst.org/neisseria/NHBA/.
Since the introduction of a meningococcal A polysaccharide conjugate vaccine MenAfriVac® in the African Meningitis Belt, outbreaks of meningitis caused by non-serogroup A meningococci, particularly W, are occurring with increased frequency. The development of a protein-based vaccine that can provide broad protection is an attractive prospect. An approach to understanding whether protein-based vaccines could have an impact on reducing the burden of meningococcal disease in the African Meningitis Belt, is to examine the genetic diversity of carriage and disease isolates of serogroup W. In this study, we focused on investigating the molecular diversity of four OMP vaccine antigens of 31 carriage and disease isolates of serogroup W from Ghana and Burkina Faso. The strains studied were isolated between 2003 and 2009 and contain conserved fHbp, porA and nadA genes, suggesting little antigenic diversification over time. A stop codon was identified among over half of the nhbA genes sequenced and was associated with a lack of expression of NHBA protein.
Previously, Pajon et al. performed a molecular characterization of 106 invasive meningococcal isolates from 13 African countries, 26 of which were from Burkina Faso and 3 from Ghana. Of the serogroups W analysed in the study, 58% were fHbp variant 2, in common with all W isolates from our collection, while 34% were variant 1 and 8% variant 3. Concordant with our findings, 98% of W were PorA subtype P1.5,2 or a related subtype indicating a marked homogeneity of PorA type among African serogroup W isolates19. A more recent longitudinal study found that a hypervirulent ST-11 serogroup W clone was responsible for most meningococcal disease in 2011 and 201224. All the isolates expressed PorA 1.5,2 and 96.4% had FetA (iron-regulated outer-membrane protein which is involved in uptake of siderophores25) variant F1-1. In accordance with our study, these two previous studies emphasise the limited diversification of major OMPs in the serogroup W meningococcal population in Africa. Studies with isogenic mutants with different expression levels of fHbp suggested that low fHbp expression contributes to resistance to anti-fHbp bactericidal activity19. It has been suggested that sparse distribution of antigens on the bacterial surface impedes cross-linking of two IgG anti-fHbp antibodies to correctly spaced epitopes26. Consequently, the antibodies cannot engage the complement protein C1q, preventing activation of the classical complement pathway. In the present study, the serogroup W isolates were found to express medium to high levels of fHbp when compared with a serogroup B strain known to express naturally relatively high levels of fHbp17, and there was no significant difference in expression between case and carrier isolates. This, together with the conservation of the fHbp ID among carrier and case isolates indicates that both carrier and case isolates could be targets of vaccine-induced anti-fHbp antibodies. NadA has emerged as an important protein for adhesion and invasion, and has been shown to elicit bactericidal antibodies8. In this study, the presence of the nadA gene in most case and carrier strains isolated from African countries suggests that NadA could be a potentially important vaccine antigen to be included in a GMMA vaccine for Africa.
This longitudinal study of meningococcal serogroup W isolates from two African countries, together with the findings of other studies, suggests that there is limited antigenic variation of meningococcal outer membrane proteins that induce bactericidal antibodies. These findings support a strategy of using protein-based vaccines, such as GMMA, to prevent meningococcal meningitis in Africa caused by serogroup W.
F1000Research: Dataset 1. Data of fHbp and NadA expression in serogroup W isolates from Ghana and Burkina Faso, 10.5256/f1000research.3881.d3632628
OK and CAM conceived the study. OK designed the experiments. EI carried out the research. EI, OK and CAM wrote the manuscript. AH, AS and GP provided the group W isolates studied. All authors were involved in the reviewing of the manuscript and have agreed to its final content.
OK and CAM are both employees of the Novartis Vaccines Institute for Global Health. CAM has received grant support from GlaxoSmithKline.
This work was supported by an EU FP7 Marie Curie Actions Industry Academia Partnerships and Pathways (IAPP) Consortium Programme, entitled GENDRIVAX (Genome-driven vaccine development for bacterial infections).
We would like to thank Dan Granoff for providing the monoclonal antibody JAR31 used in this study.
Supplemental file. Sequencing analysis of fHbp, porA, nhbA and nadA genes from serogroup W strains.
The genes encode fHbp v.2, ID 22 or 23, PorA subtype P1.5,2, NHBA allele 17 and NadA allele 3 with and without insertion sequence IS1301. DNA sequences were translated into protein sequences and sequence queries were performed using the database on http://pubmlst.org/neisseria.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: KLS was previously an employee of Novartis Vaccines.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 03 Nov 14 |
read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)