Keywords
Actinobacteria, Biotechnology, Natural products, Enzyme production, Jerusalem.
Actinobacteria, Biotechnology, Natural products, Enzyme production, Jerusalem.
Actinobacteria represent one of the most diverse groups of filamentous bacteria capable of surviving in a number of ecological niches due to their bioactive potential. They are representative of terrestrial microorganisms and usually are isolated from soils. Actinobacteria have gained special importance as the most potent source of antibiotics (Kandasamy et al., 2012) and other bioactive secondary metabolites (Solecka et al., 2012). Their metabolic potential offers a strong area of research. Accordingly, the role of actinomycetes in biotechnology and medicine is well known and these industries are always looking for novelty bioactive compounds. While most of the studies on actinobacteria have focused on antibiotic production, only few reports have focused on their enzymatic potential.
Actinobacteria are considered as a promising source of a wide range of enzymes. Some of them are produced on an industrial scale, but many other remained to be harnessed. The bacteria have the ability to degrade a wide range of hydrocarbons, pesticides, and aliphatic and aromatic compounds (Sambasiva Rao et al., 2012). They perform microbial transformations of organic compounds, a field of great commercial value. Members of many genera of actinobacteria have potential for use in the bioconversion of underutilized agricultural and urban wastes into high-value chemical products (Crawford, 1988). Some actinobacteria secrete enzymes responsible for the degradation of lignocelluloses in lignin, cellulose and hemicellulase, others may secrete enzymes that can only partially achieve this breakdown (Mason et al., 2001). Here the purpose of this preliminary study was to isolate and screen new actinobacterial isolates for their ability to degrade organic compounds via the secretion of enzymes like amylase, cellulase, protease, tyrosinase, lipase, catalase, and phosphatase. Due to the high disturbance level of biodeterioration occurred in the area of Al-Aqsa mosque in Jerusalem, Israel, the soil specimens from such place may contain novel actinobacterial colonies. Meanwhile, to the author’s knowledge, no previous studies concerned organisms involved in biodeterioration in this location.
Soil samples were collected 5 cm below the soil surface from two different sites at Northern part of Al-Aqsa mosque in Jerusalem. The soils collected from the area around Al-Aqsa are characterized by high pH ranged from 8.15 to 8.32 with organic carbon 9.61% – 12.28%. Soil textures were clay and clay loam. No attempts have been made before to isolate actinomycetes from these areas. The soil samples collected were subjected to sieving to remove plant debris and were then pre-treated by drying in open air for 2 days. Samples of 5 g were mixed with 50 ml of sterile saline solution (0.85% NaCl) and incubated at room temperature (27±2°C) for 1 hour on orbital shaker with vigorous shaking. Soil suspension was subjected to serial dilutions and then pipetted and spread onto starch casein medium (SCM, with the following ingredients gm-1Le: casein powder 1.0, starch 10.0, sodium nitrate, 3.0; agar 15.0; final pH 7.2±0.2) supplemented with antifungal cycloheximide (50 mg-1L) as described by Mansour (2003). After 7 days of incubation at 30°C, actinomycete colonies were picked up and purified onto SCM medium using streak plate techniques. The pure colonies of actinomycetes were subcultured onto starch casein slants and incubated for 3–7 days at 30°C.
Purified actinobacterial isolates were identified to genus level using different tools, morphological, cultural and physiological characteristics following the standard techniques as presented in Bergey’s Manual of Systematic Bacteriology (Anon, 1989). For morphological characterization, sterile slide oblique technique was applied (Mansour, 2003). Actinomycete colonies were streaked onto SCM, where the slide was inserted in the agar plate with an angle of 45° and incubated at 30°C for 3 and 7 days. After each incubation period, the growth of actinomycetes was examined taking the slides out from the agar and staining the actinobacteria growth using Gram stain. The slides were then examined using a light microscope (Leica, Model DMLB). Spore orientation and their morphological types were examined. Culture characterization was carried out using different culture media: SCM (Kuster, 1959), glycerol asparagine agar (reagents g-1L: L-asparagine, 1.0; dipotassium phosphate, 1.0; trace salt solution (ml) 1.0; agar 20.0; 1ml of trace salt solution contains, ferrous sulfate heptahydrate, 0.001; manganese chloride tetrahydrate, 0.001; zinc sulfate, heptahydrate 0.001; final pH 7.4±0.2 (Pridham & Lyons, 1961), glucose asparagine agar (reagents g-1L: glucose, 10.0; asparagine, 0.5; di-potassium hydrogen phosphate, 0.5; agar, 15.0; pH 7.4 (Waksman, 1961), yeast extract-malt extract agar (reagents g-1L: yeast extract, 3.0; malt extract, 3.0; dextrose, 4.0; agar, 20.0; pH 7.2 (Pridham et al., 1956), inorganic salt-starch agar (reagents g-1L: soluble starch, 10.0; di-potassium hydrogen phosphate, 1.0; magnesium sulfate, 1.0; sodium chloride, 1.0; ammonium sulfate, 2.0; calcium carbonate, 2.0; trace slats solution, 1 ml; agar, 15.0 (Kuster, 1959) and oat meal agar (reagents g-1L: oat meal, 2.0 and agar, 15.0 (Kuster, 1959). All chemicals used are from Sigma Company. The color of aerial and substrate mycelia grown on the different media used were recorded in addition to pigment production. Carbon utilization, nitrate reduction, melanin production, gelatin liquefaction and H2S production (Kuster & Williams, 1964) tests were used for physiological characterization.
In order to detect the production of extracellular hydrolases, different enzymatic agar plate assays were performed. The different assays are described below.
The starch agar medium was used to detect the amylase activity (Haritha et al., 2010). The assay medium inoculated with each isolates was incubated at 30°C for 72 hours. After incubation, the amylolytic activity was detected by flooding the agar plates with Gram’s iodine solution (2.0%). The change in color of clear zones around the growing colonies to dark blue was considered as positive.
Cellulase production was performed in agar plates supplemented with carboxymethyl cellulose (CMC) (0.5%) as the only carbon substrate, after incubation at 30°C for 72 hours. Three replicates were used for the each actinomycete isolates. The plates were then flooded with Congo red and NaCl. The yellow zones around colonies in respect to the red background indicated positive cellulose activity (Rathnan & Ambili, 2011).
The relative activity of protease production was detected for actinomycete isolates on milk agar plate, containing basal salt of starch casein amended with 20% of skimmed milk, following the method of Jani et al., (2012). The actinomycetes were grown in the middle of the milk agar plate and incubated for 5–6 days and at an interval of 24 hours. Zones of casein hydrolysis (clear zones) indicated positive results.
Tyrosinase activity was assessed in medium containing L-tyrosine (Sambasiva Rao et al., 2012). Plates containing L-tyrosine were inoculated with each tested isolate separately and then incubated at 37°C for 72 hours. The appearance of black or brown color around the margin of colonies and diffused to the medium indicated tyrosinase activity.
To observe lipase production, the actinomycetes were grown on modified medium of Vishnupriya et al., (2010) in which tween-20 (0.5%) was used instead of olive oil. Agar plates were inoculated and incubated at 30°C for 72 hours. The clearance zone around colonies was considered a positive evidence of tween-20 hydrolysis.
All isolates obtained were screened for catalase activity after 3 to 4 days of subculture on newly fresh SCM following the method of Mahon et al., (2011) using the slide (drop) method. Positive reactions were evident by immediate effervescence (bubble formation).
Acid and alkaline phosphatase activities were determined according to Ghorbani-Nasrabadi et al., (2013). Inoculated medium supplemented with CaHPO4 (5 g/l) were incubated at 37°C for 48 to 72 hours. Phosphatase active isolates were recorded based on the halo-zones produced around the colonies.
Evaluation of enzymatic activity. The enzymatic activity (EA) of different tested substrates was examined. The diameter of growth was measured and the clear zone representing enzyme activity was calculated by using the formula:
EA = Diameter of zone of tested substrate hydrolysis - Diameter of colony in cm.
Based on the EA test, the organisms can be categorized into three groups: showing excellent activity (EA>2), good (EA<2) and poor (EA<1).
A total of 35 actinobacteria isolates were obtained from the two different soils collected from Northern part of Al-Aqsa mosque in Jerusalem. From Site 1, only 17 isolates were recovered in which four genera, Actinomadura, Streptomyces, Elylrosporangium and Actinopolyspora, were represented (Table 1). Site 2 was represented by more diverse genera of 18 isolates, Streptosporangium, Actinomadura, Nocardiopsis, Nocardia, Elytrosporangium and Actinopolyspora (Table 1). Genus Actinomadura was represented with the highest frequency in both sites (52.95% and 33.33% for site 1 and site 2 respectively). Despite the fact that isolation methods reveal only a minor fraction of the real existing microbes (Groth et al., 1999) we could demonstrate a great diversity among actinobacteria in the studied sites. To our knowledge (Thaer et al., 2009), this is the first study reporting identification of actinomycetes from this terrestrial environment. Meanwhile, the genera obtained are the first to be recorded in Jerusalem or in the whole country.
The differences observed among the genera of actinobacteria identified in both sites studied, may be due to the different human activities, including construction in such area. These differences may also indicate that the methods proposed and employed for actinobacteria isolation may not the suitable for site 1, and that soil pretreatment should be sought to explore the other genera inhabiting the area. Machavariani and colleagues (2011) found that preliminary treatment of soil by chemical substances like adrenaline and heteroauxin exerts a positive influence on the germination of actinomycete spores and contributes to the natural product activity of the isolated strains. Moreover, they demonstrated that soil pretreatment with different chemical substances play a role for the most complete isolation of actinomycetes that inhabit certain soil.
When screening enzyme activities, 32 of the 35 actinobacteria isolate showed a good amylolytic activity (Figure 1A) and the other three isolates (site 2), Streptosporangium sp.2, Nocardia sp.2 and Elylrosporangium sp.4, had a moderate activity (Figure 1B). However, all isolates from site 1 had a good amylase activity (Table 1). For other enzymes, although actinomycetes are well known as potent degraders of cellulose, lignin, chitin and other complex polysaccharides (El-Fiky et al., 2003; McCarthy & Broda, 1984; Prasad et al., 2012; Wilson, 1992) none of our isolates were able to produce cellulase enzyme. Our results seem to be in contrast with previous studies, and this may be explained by the unsuitable culture conditions for cellulase production such as optimal pH as reported by (Rathnan & Ambili, 2011). In their study, they showed that cellulase enzyme production by Streptomyces sp. using fruit waste as substrate was the highest at alkaline pH.
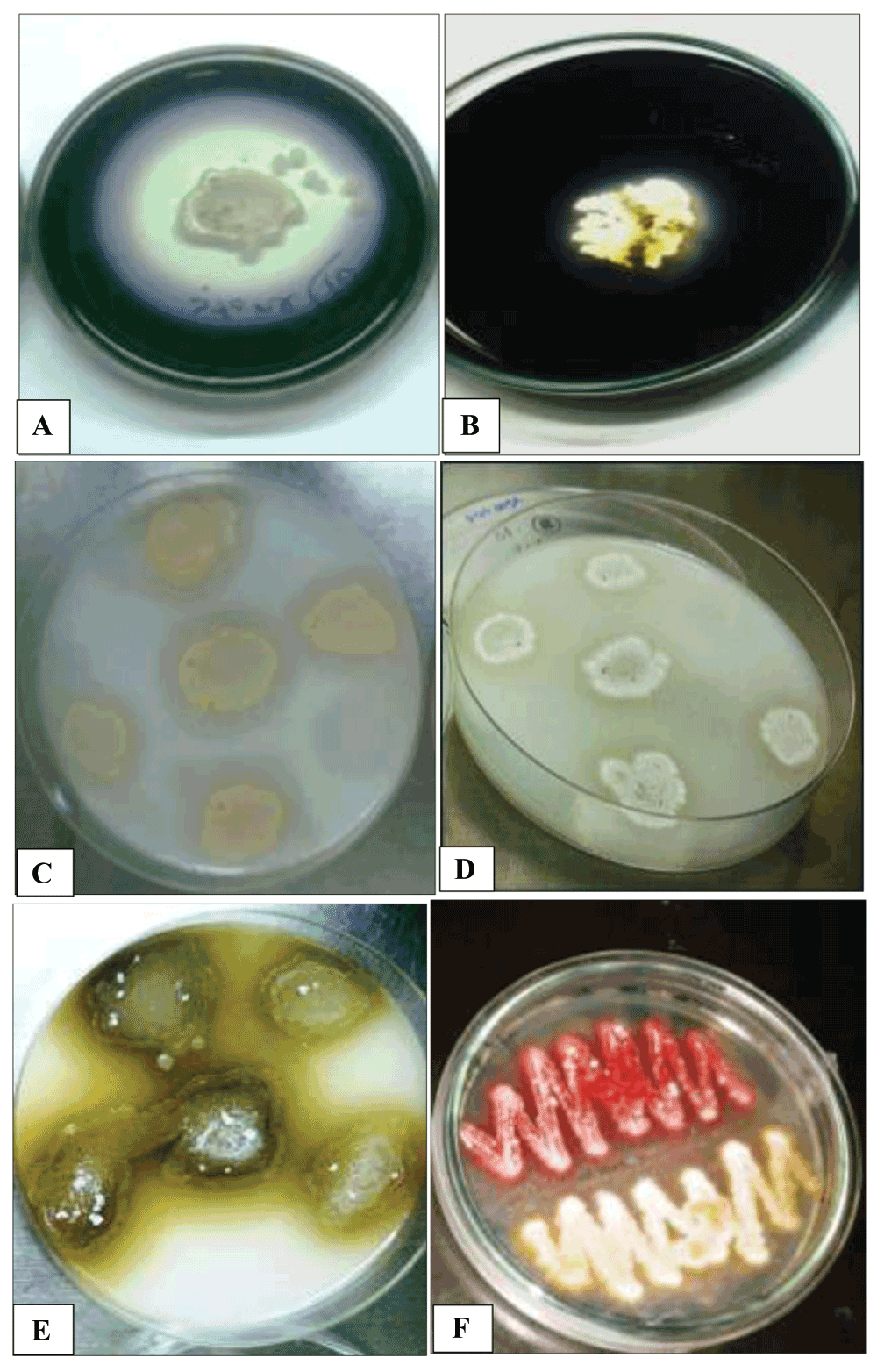
(A and B) showing good to moderate amylase activity respectively; (C and D) showing moderate protease activity indicated by clear zones around colonies; (E) showing good tyrosinase activity indicated by deep brown pigments and (F) showing no activity for two different actinomycete isolates.
Proteases represent one of the most important groups of enzymes and have been shown to play a role in many industrial and medical fields (Prakash et al., 2013). They can be used in detergent, food pharmaceutical, leather, waste processing industries and silk industries. Proteolytic enzymes have already been used in various forms of therapy and their use in medicine is gaining interest. Therefore, searching for new actinomycete strains still is the focus of many studies (Prakash et al., 2013). Our results revealed that only three isolates, Streptosporanium sp.1, Streptosporangium sp.2 and Actinomadura sp.10 were able to produce protease enzyme (Figure 1C and D). These three isolates were obtained from site 2. However, the site 1 and the reset of isolates from site 2 completely failed to show any protease activity. This rather low estimate of active strains may indicate that the method used for preliminary screening is not accurate (Jani et al., 2012). In addition, the low activity observed may suggest that our culture conditions are not optimal for such test (Jani et al., 2012).
The search for novel tyrosinases is still in need due to their potential in industrial applications and medical purposes. Tyrosinases have been suggested as potential tools in treating melanoma and as potential antioxidants, antiviral agents and immunogens (Popa & Bahrim, 2011). Among the isolates, only few showed tyrosine activity but all of these exhibited strong activity (Table 1). Only one isolate out of 17 (5.9%) in site 1, Elylrosporangium sp.3, showed high production of tyrosinase and the rest failed to show any activity (Figure 1E and F). Meanwhile, four isolates out of 18 (22.2%) of site 2, Streptosporangium sp.2, Kitasatosporia, Nocardia sp.2, and Actinomadura sp.15 showed different tyrosinase activity. These isolates had a high potential for enzyme production except Nocardia sp.2 which showed moderate activity.
For lipase production, all isolates from site 1 failed to show any activity. However, three isolates (16.7%) out of 18 isolates of site 2, Nocardiopsis sp.1, Nocardiopsis sp.2 and Kitasatosporia sp., were able to produce lipase in a weak to moderate manner of activity (Table 1).
Catalases are ubiquitous enzymes and have been isolated from a broad range of procaryotic and eukaryotic organisms (Zámocký et al., 2012). Actinobacteria are aerobic bacteria and would be expected to have catalase activity; however, our actinomycete isolates recovered from both studied sites showed different ranges of positive activity (Figure 2). The catalase activity was observed at pH 7 of the growing medium for all isolates. Out of the isolated actinomycetes, Kitasatosporia sp. was the most active catalase producer, followed by 42.9% of the isolates have moderate catalase activity (Table 1). However, 18 isolates (51.4%) including different genera showed weak activity. The low expressed activity may be due to shortage of manganese ions or iron concentration in the growing medium, since catalases rely on iron or manganese for their activity (Mishra & Imlay, 2012). Further studies are needed to identify the factors that weaken the scavenger process.

A and B show the activity of isolates from site 1 and site 2 respectively. Evolved oxygen bubbles indicated a very strong activity and weak bubbles indicated a weak activity.
Since phosphate always exists in unavailable form for plant growth, phosphatase activity is an important mechanism of solubilizing inorganic phosphate (Sharma et al., 2013). In our study, all strains listed in Table 1, grown at room temperature (approximately 27±2°C) on basal medium supplemented with CaHPO4 (5 g/l) were tested for phosphatase activity at pH 7.0. Among the isolates, only Streptosporangium sp.1 and Nocardiopsis sp.2 showed phosphatase activity. The failure to detect of phosphatase activity by the remaining isolates may be due to the medium composition (Fredrikson et al., 2002). In a recent study done by Ghorbani-Nasrabadi et al., (2013), it was shown that substitution of nitrogen source in the growth medium by organic or inorganic nitrogen sources resulted in a significant reduction of phosphatase activity.
In conclusion, the need for low cost enzymes that can be applied in diverse biotechnological industries could be satisfied with the discovery of novel enzymes and metabolites. Moreover, the application of genetic engineering techniques in enzyme manufacturing is dramatically sparking the exploitation of new enzymes and the development of new enzyme properties. Actinobacteria have been proved a reservoir of important enzymes and metabolites due to their versatile genetic repertory (Prakash et al., 2013). Identification of new actinobacterial isolates in unique ecological environments could yield molecules that could become future players in green technology. Therefore our study contributes to explore new ecological sites for actinobacterial identification. Actinobacterial isolates that inhabitant the northern part of Al-Aqsa Mosque, showed a diverse population in the two sites where the soil was collected. These genera have been identified in each site for the first time. However more suitable growth conditions should be tested to explore the metabolites and enzymatic activities of these organisms.
Mansour S., Abedelazeium A. and Deraz S. conceived the study. MS and AA designed the experiments. MS and DS carried out the research. AA and MS provided the expertise throughout the experiment. MS prepared the first draft of the manuscript. All authors were involved in the revision of the draft manuscript and have agreed to the final content.
The authors would like to thank Prof. Louis Tisa for revising the manuscript. Also thanks go to the researchers A. Salh and A. Hamza for their help during the lab work.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
References
1. Kim CJ, Lee KH, Shimazu A, Kwon OS, et al.: Isolation of rare actinomycetes on various types of soil. Kor. J. Appl. Microbiol. Biotechnol. 1995; 23: 36-42 Reference SourceCompeting Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 14 Jan 15 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)