Keywords
Fluorescence microscopy, telomeres, telomere elongation, telomere spatial organization, time-lapse microscopy, single-molecule fluorescence, chromosome
Fluorescence microscopy, telomeres, telomere elongation, telomere spatial organization, time-lapse microscopy, single-molecule fluorescence, chromosome
Oskar Heimstädt, who built the first fluorescence microscope, ended his 1911 paper with the following perspective: “If and to what degree fluorescence microscopy will widen the possibilities of microscopic imaging only the future will show”1. More than a century later, fluorescence microscopy has proven transformative in our ability to illuminate almost all aspects of cellular biology. One of the more recent frontiers in fluorescence microscopy is the resolution of biological phenomena at the single molecule level, called single molecule fluorescence2. Biological processes have evolved to be inherently heterogeneous, transient and dynamic3, and therefore difficult to track. Single molecule microscopy often permits a birds-eye view of ephemeral and complex mechanisms. This review will focus on selected recent advances in high-resolution microscopy including, but not limited to, single-molecule fluorescence microscopy, that have enlightened our understanding of chromosome ends and the enzyme that replenishes them.
Many organisms must contend with the vulnerability of linear chromosome ends to enzymes that degrade, rearrange, or incompletely replicate DNA. This susceptibility to breakage, and their distinct “knob-like” appearance under the light microscope, made them an early target of study by scientists such as Barbara McClintock and Hermann Müller4. We now appreciate that telomeres are a highly specialized nucleoprotein structure whose maintenance is critical to genome stability (Figure 1A–C)5,6. Replenishment of the G-rich sequences that comprise the telomeres is carried out by telomerase, whose core components are a reverse transcriptase and an integral RNA that provides the telomere template (Figure 1D)7.
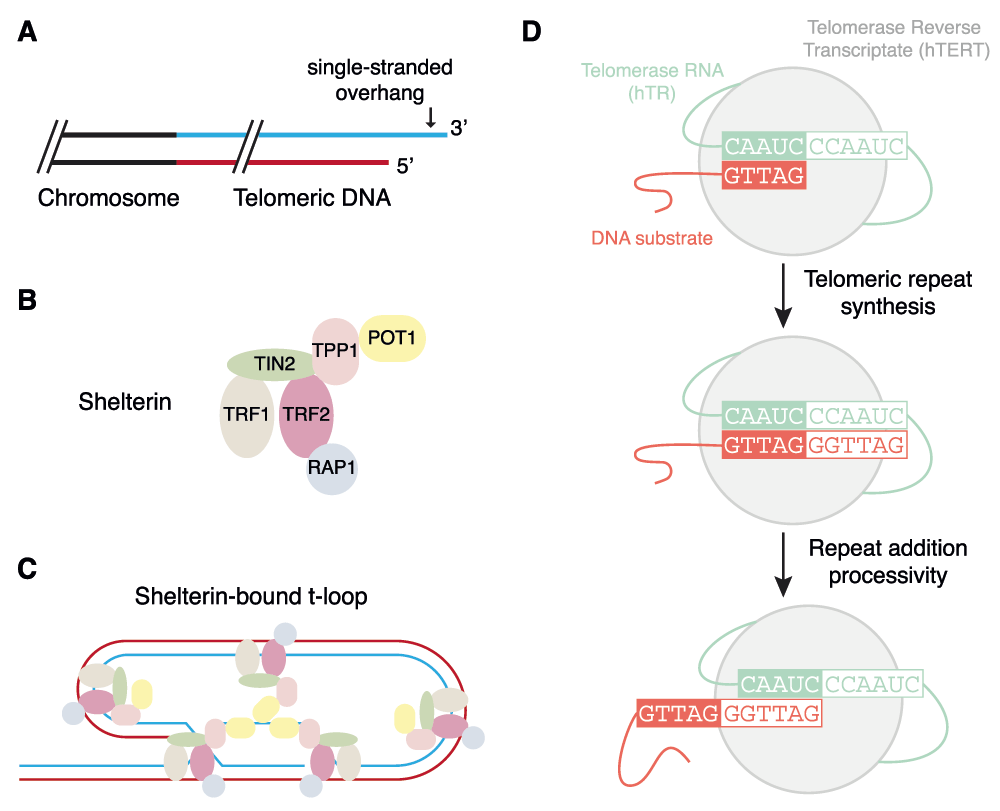
(A) In many organisms, chromosome ends terminate in a single-stranded, G-rich overhang preceded by up to several kilobase pairs of double-stranded G-rich DNA. (B) Telomeres are capped by a six-subunit complex called shelterin. (C) Shelterin (particularly TRF2) promotes the formation of a higher order telomeric loop (T-loop) structure that serves to mask telomeres from the deleterious fates associated with a free DNA end5,6. (D) The catalytic cycle of the core telomerase enzyme, comprised of a protein (TERT) and RNA (hTR)7.
Fluorescence microscopy has revolutionized our ability to probe the length, location, and recombination of telomeres in vivo through the application of fluorescently labeled peptide nucleic acids that bind tightly and specifically to telomeric DNA8–10. These techniques have uncovered interesting distinctions in the three-dimensional (3D) localization of mammalian telomeres in normal cells versus cancer cells11,12. Time-lapse microscopy has also revealed increased mobility of telomeres upon induction of a DNA break13 as well as unexpected long-range telomere associations in telomerase-negative cells after DNA breakage14. In the budding yeast Saccharomyces cerevisiae, fluorescent tagging of several components of telomerase have also provided considerable insight into the temporal and spatial dynamics of telomerase recruitment in living cells15–18.
Fluorescence microscopy is also being combined with other leading-edge technologies to directly target specific genomic regions, including the telomere. Using the CRISPR/Cas system in which Cas9 can be guided to specific genomic loci using a small RNA19, Chen and colleagues targeted an enhanced green fluorescent protein (EGFP)-tagged catalytically inert Cas9 specifically to telomeres20. Telomere-recruited Cas9 demonstrated a punctate pattern that spatially overlapped with that of the telomeric DNA binding protein, telomeric-repeat binding factor 2 (TRF2). Furthermore, EGFP intensity correlated linearly with telomere intensities obtained using fluorescence in situ hybridization (FISH) (Figure 2A)20. This method could also be employed to visualize Cas9 directed to a single genomic locus20. Technological advances such as these may soon permit the ability to explore the spatial and dynamic localization of telomeres and telomerase in living mammalian cells to a similar extent as has been explored in budding yeast.
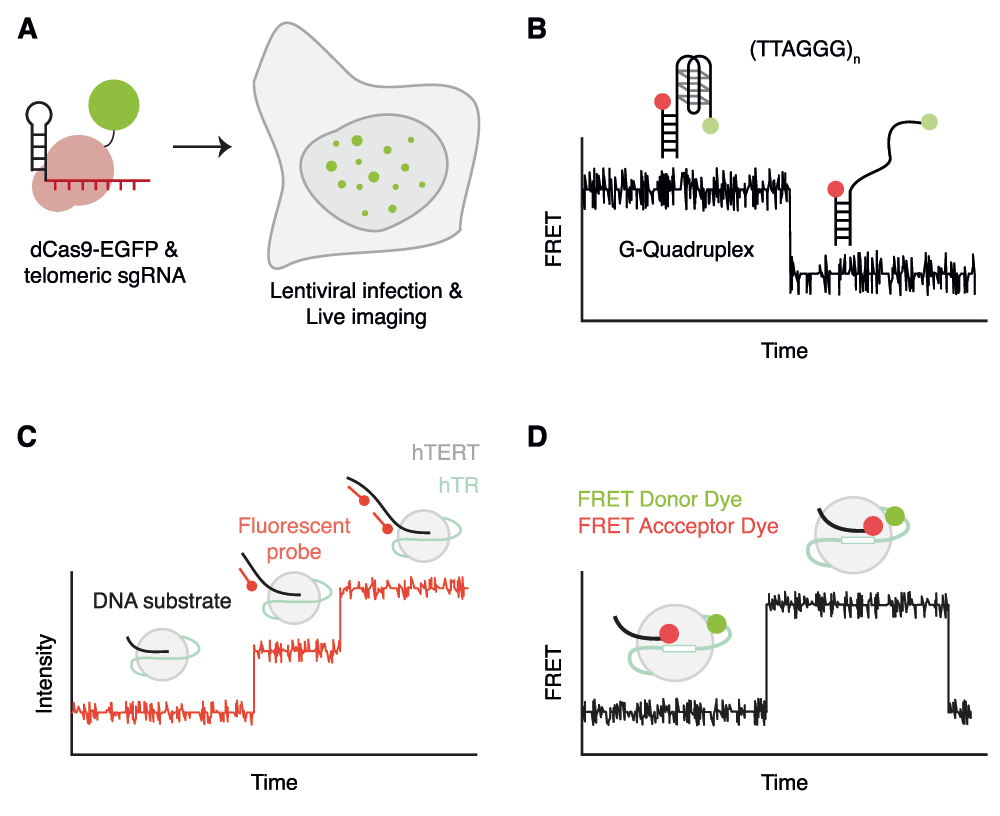
(A) The use of a fluorescently tagged Cas9 and a guide RNA specific to telomeres to measure telomere dynamics and length in live cells20. (B) The ability of single-molecule Förster resonance energy transfer (smFRET) to measure the dynamics of G-quadruplex folding27,30–36,38. (C) The application of fluorescent probes complementary to telomeric substrates to measure the elongation properties of telomerase47. (D) The application of FRET to assess the intermolecular proximity of the DNA substrate and the RNA subunit of telomerase during telomere synthesis48.
The telomere terminus ends in a 3’ G-rich overhang of variable length that can invade the upstream double-stranded telomeric DNA to form a structure called a T-loop21. Super-resolution microscopy, specifically stochastic optical reconstruction microscopy (STORM), has recently enabled an unprecedented visualization of T-loops that are crosslinked and purified from murine cells22 (Figure 1B,C). The loss of T-loops occurred specifically upon loss of the shelterin subunit TRF2, and not other shelterin components, which provides an elegant demonstration that it is the T-loop structure that protects ends from the deleterious molecular events observed when TRF2 is removed from telomeres22,23.
Telomeric DNA sequences can also form in vitro intra- or inter-parallel structures called G-quadruplexes, and evidence is accruing to suggest their existence at telomeres and at interstitial G-rich regions24. Single-molecule Förster resonance energy transfer (smFRET) has been applied to the molecular dynamics of G-quadruplex formation in vitro25–28 (Figure 2B). The smFRET approach has several advantages that permit a high-resolution view of G-quadruplex dynamics. Firstly, this technique can resolve different conformations using FRET signal intensities and Gaussian curve fitting29. Secondly, individual single-molecule traces reveal dynamic switching between states in real time. Hidden Markov modeling can then extract the dwell time of each molecule, which is a reflection of the stability of each state29. Applying smFRET to the analysis of shelterin binding to telomeric substrates in vitro, several groups have investigated the intricate relationship between G-quadruplexes and POT1/TPP1 (protection of telomere 1/Pot1-interacting protein TINT1-PTOP-PIP1)30,31. Furthermore, this technique also revealed the binding and unfolding of G-quadruplexes by proteins such as RAD51, WRN, BLM, RecQ and RPA32–36. Recently, the use of smFRET combined with magnetic tweezers spectroscopy37 has been used to measure the thermodynamic properties of G-quadruplex folding38. These findings provide important insights into the dynamics of telomeric DNA structure in a more native nucleoprotein context.
The telomerase reverse transcriptase, TERT, is able to synthesize new telomeric DNA, one nucleotide at a time, by virtue of an integral telomerase RNA that contains a short telomere-complementary sequence (Figure 1D). Although TERT shares several features in common with other viral reverse transcriptases, one unique aspect is its ability to repeatedly copy the same template for many cycles in an iterative process termed repeat addition processivity (RAP)39,40. Although much information has been gleaned using standard biochemical techniques regarding the complex DNA-protein, RNA-protein and protein-protein interactions that contribute to telomerase RAP (for a few current examples, see 41–46), it is only recently that single-molecule fluorescence has been brought to bear on the elongation properties of telomerase in vitro47. Hwang and colleagues immobilized immunopurified telomerase from cell extracts on a surface and employed a fluorescently labeled probe complementary to telomeric DNA to obtain a digital readout of telomerase activity in real time (Figure 2C). Their results suggested that active telomerase can exist in two dynamic states defined as an initial activation period followed by an extension period in which telomere elongation was visualized as a step-wise increase in fluorescence47. In addition, they found that TPP1-POT1 stimulated the elongation rate and overall product length47, consistent with previously described properties of TPP1-POT1. Although the nature of the fluorescent probe binding (which comprises the sequence 5′-CCCTAACCCTAACCC-3′) precludes single base resolution, this technique promises to provide an unprecedented, single-molecule view of the intricacies of the telomerase elongation cycle.
In another smFRET approach, the distance between the region 5’ of the RNA template of telomerase and a DNA substrate was followed during telomeric repeat synthesis (Figure 2D)48. Using different combinations of regular and chain-terminating nucleotides, information about the position of the DNA oligonucleotide substrate was registered at single-base pair resolution during elongation. While the substrate appears in a compact conformation after initial binding by telomerase, after a round of DNA synthesis the substrate realigns with the template via Watson-Crick base pair interactions. The aforementioned study by Parks and colleagues suggests that DNA dissociation and realignment is not the limiting step, and that an additional conformational adjustment is necessary after DNA:RNA repositioning to re-acquire a catalytically competent state48. This study illustrates the power of non-linear Gaussian curve fitting and hidden Markov modeling to extract the dwell time of the different FRET states from individual traces, which in turn permits a very precise dissection of the catalytic mechanism, albeit only over the short distances in which FRET can be observed.
smFRET has also been used to evince the real-time dynamics of folding of the pseudoknot domain with the telomerase RNA49–51. It has also been applied to examine the assembly and activity of the Tetrahymena thermophila telomerase RNP51–53 and, more recently, the role of the N-terminal domain of T. thermophila TERT in the stabilization of short RNA:DNA hybrids during telomerase catalysis54. In addition to valuable insights into telomerase catalysis, these techniques might also permit a precise elucidation of the mechanism-of-action of chemical modulators of telomerase activity, as well as an in vivo determination of human telomerase component stoichiometry, as was recently demonstrated for budding yeast telomerase18.
In the year in which fluorescence microscopy was first described, Arthur Brisbane offered the sage advice: “Use a picture. It’s worth a thousand words.”55. We have reached a technological watershed in biology that will enable an unprecedented single-molecule and high-resolution view of the inner workings of many cellular machines. As we have illustrated here with but a few selected examples, fluorescence microscopy can be applied in many different ways to different problems, but key advances are the ability to dissect individual events instead of ensemble, population-based outputs, and to permit dynamic measurements in living cells, in real time. How far we have come from seeing telomeres as a cytogenetic “knob”, and how far we have yet to come.
We thank Paul Maddox, Tracy Bryan, Michael Stone, and Raymund Wellinger for comments and constructive input on the review.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 11 Dec 15 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)