Keywords
psoriasis, delayed-type hypersensitivity, immune regulation, diphencyprone
psoriasis, delayed-type hypersensitivity, immune regulation, diphencyprone
Diphencyprone (DPCP) is a hapten that induces delayed-type hypersensitivity (DTH) reactions in human skin, and is used therapeutically for alopecia areata1, warts2, and cutaneous melanoma metastases3. The mechanisms by which DPCP decreases pathogenic immunity for the promotion of hair growth in alopecia areata are incompletely understood. DPCP has been shown to alter the cytokine profile in treated alopecic scalp, in particular increasing interleukin (IL)-2 and IL-10 expression4. This increased IL-10 expression has been hypothesized to inhibit the lesional T cells of alopecia areata, but a comprehensive evaluation of other negative immune regulators induced by DPCP is lacking. We have previously shown that human skin responses to DPCP evolve from an inflammatory/effector peak at 3 days post-challenge to a more regulated immune response, with diminished markers of T cell activation, at 14 days. This study included comprehensive gene expression profiling, by microarray and qRT-PCR approaches, of biopsies from DPCP-challenged healthy volunteer skin at 3 days (peak reaction), 14 days (actively resolving reaction), and 120 days (4–8 months; fully resolved reaction) compared to placebo-treated skin5. We have also previously performed similar transcriptomic profiling of psoriasis vulgaris lesional vs. non-lesional skin. This resulted in a meta-analysis derived transcriptome (MAD3) which combined the results of 3 individual microarray experiments, in an effort to address the variability in differentially expressed genes observed between experiments6. In this study, we expand our previous characterizations of transient DTH reactions and chronic psoriasis biopsies by directly comparing them to each other, particularly in relation to positive and negative immune regulation.
For diphencyprone (DPCP) reaction microarray, qRT-PCR, and immunohistochemistry studies, skin biopsies were obtained from 11 volunteers under a protocol approved by The Rockefeller University’s Institutional Review Board (IRB Number JKR-0742). Written, informed consent was obtained from all subjects and the study adhered to the Declaration of Helsinki principles. This trial is registered at clinicaltrials.gov under NCT01452594 (https://clinicaltrials.gov/ct2/show/NCT01452594). For each volunteer, biopsies were taken of placebo-treated skin as well as DPCP reactions 3, 14, and 120 days after challenge, as previously described5.
For psoriatic lesional vs. non-lesional skin microarray data, we used the meta-analysis derived (MAD3) transcriptome described in 6. Psoriatic lesional tissue for qRT-PCR and immunohistochemistry studies were from deidentified residual samples of plaque-type psoriasis vulgaris from previous studies for whom no clinical characteristics are available; a psoriasis area severity index of more than 12 (moderate-to-severe psoriasis vulgaris with >10% body surface area involvement) was required for entry into these trials.
Total RNA was extracted using the miRNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol with on-column DNase digestion. The amount of RNA was assessed by NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE). The quality of extracted RNA was examined using Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA). RNA was hybridized to HGU133 Plus 2.0 chips (Affymetrix, Santa Clara, CA) to measure relative gene expression.
Statistical analysis. Microarray data were analyzed using R/Bioconductor packages (http://www.r-project.org). The Harshlight package7 was used to scan Affymetrix chips for spatial artifacts. Expression values were normalized using the GeneChip Robust Multi-array Average (GCRMA) algorithm. Genes with low variation and low expression in most samples were filtered out prior to the analysis. Batch effect due to hybridization date was adjusted using ComBat8. To identify differentially expressed genes, we fitted by REML (Restricted Maximum Likelihood) a linear mixed effect model with treatment (placebo/DPCP) and day (3/14) as fixed effects and a random intercept for each patient. Hypotheses of interest were tested using contrasts in R’s limma package framework. The p-values resultant from the moderated paired Student’s t-tests were adjusted for multiple hypotheses using the Benjamini-Hochberg procedure, which controls for the false discovery rate. The DPCP data discussed in this publication have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GSE accession number GSE52360, http://www.ncbi.nlm.nih.gov/geo/). Psoriasis data were derived from6.
Pre-amplification quantitative RT-PCR technique was used for measuring various genes in total RNA extracted from skin biopsy samples according to the company’s instructions. Briefly, 5 ng of total RNA was subjected to first-strand cDNA synthesis using High Capacity cDNA Reverse Transcription kits (Applied Biosystems, Carlsbad, CA). The resulting cDNA was subjected to 14 cycles of pre-amplification using TaqMan PreAmp Master Mix Kit (Applied Biosystems) with desired pooled assay mix. The Gene Amp PCR System 9700 (Applied Biosystems) was used for the pre-amplification reaction with the following thermal cycler conditions: 10 min at 95°C and 14 cycles of 15 seconds at 95°C followed by 4 min at 60°C. 12.5 μl of pre-amplified cDNA was then used for quantitative RT-PCR reaction using TaqMan Gene Expression Master Mix (Applied Biosystems). The 7900HT Fast Real-Time PCR System was used for PCR reactions, and the thermal cycler conditions were as follows: 2 minutes at 50°C, 5 minutes at 95°C, and 40 cycles of 15 seconds at 95°C followed by 60 seconds at 60°C. Data were analyzed by the Applied Biosystems PRISM 7700 software (Sequence Detection Systems, ver. 1.7) and normalized to human acidic ribosomal protein (hARP) housekeeping gene.
All assays were from Applied Biosystems and inventoried assays used in this study were as follows: IL10 (Hs00961622_m1), CTLA4 (Hs03044418_m1), PDCD1 (PD1) (Hs01550088_m1), CD274 (PDL1) (Hs01125301_m1), PDCD1LG2 (PDL2) (Hs01057777_m1), IDO1 (Hs00984148_m1), and LAG3 (Hs00158563_m1). For RPLP0/hARP, a custom primer/probe set was used (Forward: CGCTGCTGAACATGCTCAA, Reverse: TGTCGAACACCTGCTGGATG, Probe: 6-FAM-TCCCCCTTCTCCTTTGGGCTGG-TAMRA).
Frozen sections of skin biopsies were dried at room temperature and then fixed for 2 minutes in acetone. Next, the samples were blocked with 10% normal serum of the species in which the secondary antibody was made and then the samples were incubated overnight at 4°C with the appropriate primary antibody. Biotin-labeled secondary antibodies (Vector Laboratories, Burlingame, CA) were amplified with avidin-biotin complex (Vector Laboratories) and developed with chromogen 3-amino-9-ethylcarbazole (Sigma Aldrich, St. Louis, MO) to produce a red color indicative of positive staining.
Primary antibodies used in this study are as follows (all are mouse monoclonal): IL10 (Life Technologies, Clone 945A2A5, IgG1, 1:50 dilution), CD95 (FAS) (BD Biosciences, Clone DX2, IgG1, 1:50), LAG3 (Enzo Life Sciences, Clone 17B4, IgG1, 1:50), PD1 (eBioscience, Clone MIH4, IgG1, 1:50), PDL1 (eBioscience, Clone MIH1, IgG1, 1:50), PDL2 (eBioscience, Clone MIH18, IgG1, 1:100), IDO1 (LifeSpan Biosciences, Inc., Clone 10.1, IgG3, 1:100), and CTLA4 (abcam, Clone BNI3, IgG2a, 1:100).
Since DTH reactions naturally resolve, we sought to compare our DPCP biopsies (from 5) to those taken from patients with psoriasis vulgaris (from 6), a chronic T cell-mediated inflammatory disease that does not resolve and which, in many ways, represents amplifications of background immune circuits that exist in normal human skin9. To globally assess the balance of positive vs. negative immune regulators in both DPCP reactions and psoriasis using our microarray data, we developed one comprehensive gene list for positive regulatory or inflammatory genes and another gene list for negative regulatory or immunosuppressive genes (through Gene Ontology terms and literature review previously discussed in 5, Table 1 has “negative regulator” list and fold change values for DPCP day 3 and psoriasis transcriptomes, “positive regulator” list is derived from GO term 0002684 “positive regulation of immune system process” but with genes removed that are in common with GO term 0002683 “negative regulation of immune system process”). Our microarray data showed increased fold changes of many negative regulators in DPCP day 3 biopsies vs placebo-treated skin (DPCP day 3 transcriptome) compared to psoriasis lesional vs non-lesional skin (psoriasis transcriptome). For instance, CTLA4 expression was significantly increased 21.6-fold in the DPCP day 3 transcriptome, but non-significantly increased 3.7-fold in the psoriasis transcriptome. Venn diagrams show that the psoriasis transcriptome only has seven genes from the negative regulator list, while the DPCP day 3 transcriptome has 52 (Figure 1a). Although the DPCP day 3 transcriptome also has more genes from the positive regulator list than psoriasis, the odds ratio for the positive regulator list was not significantly different between these two transcriptomes. The odds ratio for the negative regulator list, however, was significantly different (Figure 1b). The altered balance between positive vs. negative regulatory transcripts in psoriasis compared to DPCP reactions can also be seen in Figure 1c which shows that DPCP transcriptomes at all time points (days 3, 14, and 120) have a higher ratio of negative to positive regulator genes than the psoriasis transcriptome in terms of expression levels for each gene set as a whole (as opposed to number of genes as indicated in the Venn diagrams). This is despite the fact that the DPCP day 3 transcriptome has comparable expression levels of the MAD3 psoriasis transcriptome genes to actual psoriasis samples, and therefore highlights the negative regulator expression that is unique to DPCP reactions.

(a) Venn diagrams showing overlap of MAD3 psoriasis transcriptome (left) and DPCP day 3 transcriptome (right) with both positive regulatory (Pos) and negative regulatory (Neg) gene lists (common gene lists applied to both transcriptomes). The percentages of the MAD3 and DPCP day 3 transcriptomes comprised of the positive regulatory gene list are 5.7% and 7.3%, respectively. On the other hand, the percentages comprised of the negative regulatory gene list are 0.7% and 1.5%, respectively. (b) Odds ratios (OR) of negative regulatory (red bars) and positive regulatory (blue bars) gene lists in DPCP day 3 and psoriasis transcriptomes. (c) Black bars represent negative regulator genes, gray bars represent positive regulator genes, and white bars represent all MAD3 psoriasis transcriptome genes. The y-axis shows log2 (fold change) of all genes in the given gene set. DPCP day 3 and MAD3 samples have comparable MAD3 transcriptome expression levels but there is a substantial difference between all DPCP time points (days 3, 14, and 120 or “late”) and MAD3 samples in terms of the relative levels of negative and positive regulator gene list expression. This is quantified as the “ratio of negative to positive regulator genes.”
To confirm some of our microarray findings, we performed qRT-PCR and found that psoriasis lesional skin biopsies have significantly lower expression of many negative immune regulators compared to peak DPCP biopsies. These regulators include lymphocyte activation gene 3 (LAG3), cytotoxic T lymphocyte-associated 4 (CTLA4), indoleamine 2,3-dioxygenase (IDO1), programmed cell death 1 (PD1), programmed cell death 1 ligand 1 (PDL1), programmed cell death 1 ligand 2 (PDL2), and IL-10 (Figure 2a). We confirmed the decreased expression of these and FAS (which by gene expression had 9.3- and 1.1-fold changes in the DPCP day 3 and psoriasis transcriptomes, respectively) at the protein level by immunohistochemistry (Figure 2b).
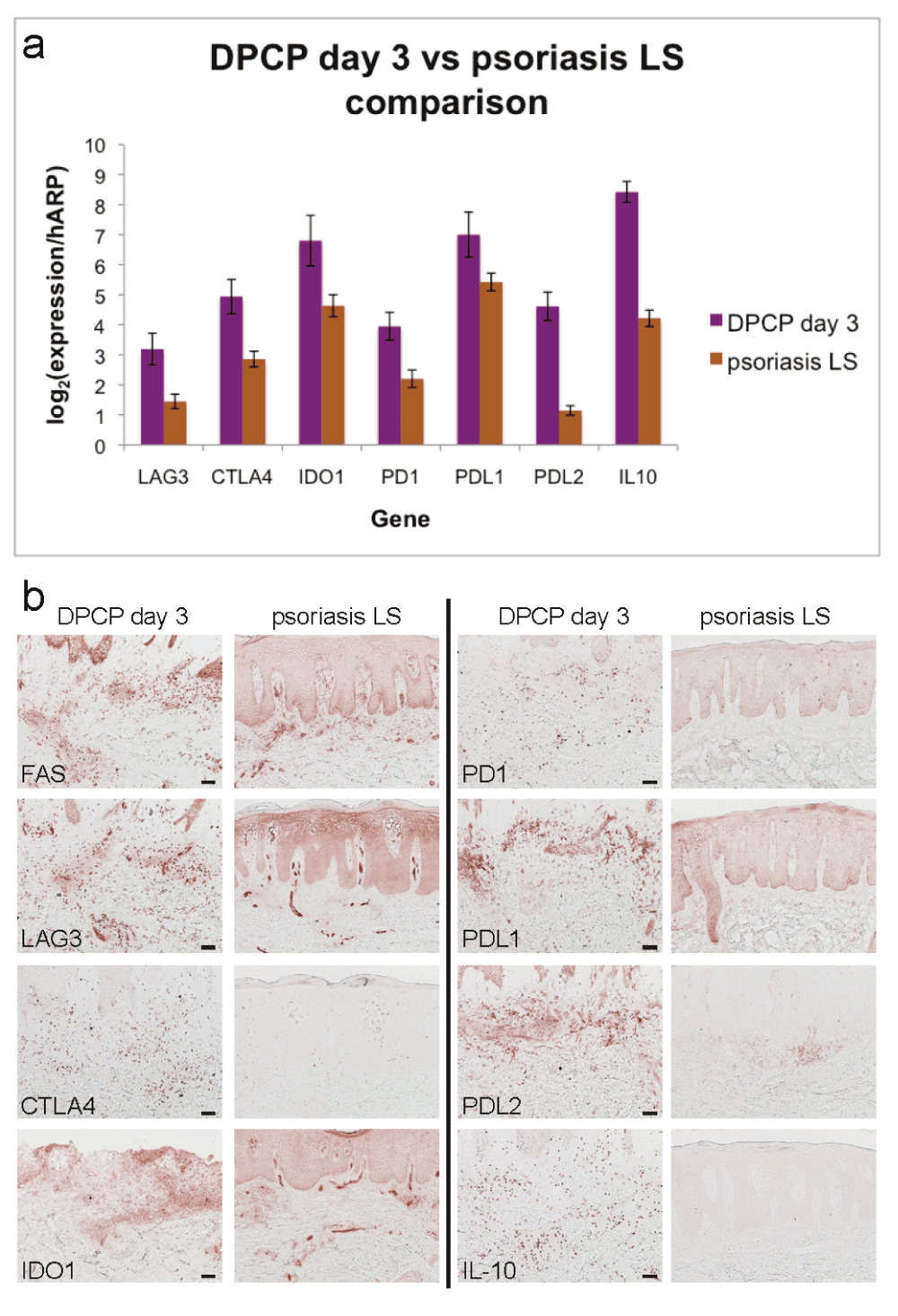
(a) qRT-PCR analysis for negative regulators LAG3, CTLA4, IDO1, PD1, PDL1, PDL2, and IL-10. Shown are average normalized expression values for DPCP day 3 samples (n=11, purple bars) and psoriasis lesional skin (LS) samples (n=11, brown bars). All except PDL1 are p<0.05 by unpaired two-tailed t-test assuming equal variance. Error bars represent standard errors of the mean. (b) Immunohistochemistry showing increased protein expression of negative regulators in DPCP day 3 samples compared to psoriasis LS. Shown are stains with antibodies specific to the indicated targets. Scale bar = 100 μm (applies to all images).
These data suggest that disease chronicity in psoriasis could be related to absence of several negative immune regulatory pathways, with the implication that strategies to obtain stable clearance/restore tolerance in skin lesions may need to focus on increasing these negative pathways. These negative immune mechanisms may be of more general importance for maintaining skin homeostasis as non-inflammatory in the presence of a large population of effector memory T cells that normally reside in skin10. In addition, these negative immune regulators are likely involved in the therapeutic applications of DPCP, particularly alopecia areata where IL-10 has already been implicated4. Further study of these regulatory mechanisms that are present in DPCP reactions, but not in psoriasis, could reveal novel factors in the pathogenesis of chronic inflammation. The DPCP system used here provides a tractable model for primary discovery of pathways potentially involved in immune regulation in peripheral tissues.
NG and JGK conceived the study and designed the experiments. NG carried out the research. MS-F and JCR contributed to the design of experiments and provided expertise in genomics. NG prepared the first draft of the manuscript. All authors were involved in the revision of the draft manuscript and have agreed to the final content.
This research was supported by National Institutes of Health (NIH) grant UL1 RR024143 from the National Center for Research Resources and the Milstein Medical Program. NG was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the NIH under award number T32GM07739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. The content of this study is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
|
Version 1 11 Jun 15 |
read | read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)