Keywords
Active compounds, X-ray structures, activity cliffs, data mining, structure-activity relationships, computational methods
This article is included in the Cheminformatics gateway.
This article is included in the Data: Use and Reuse collection.
Active compounds, X-ray structures, activity cliffs, data mining, structure-activity relationships, computational methods
The activity cliff concept has experienced increasing interest in chemical informatics and medicinal chemistry1–5. A consensus definition of activity cliffs1–4 refers to pairs or groups of structurally similar or analogous active compounds with large differences in potency4,5. For the definition of activity cliffs, the specification of similarity and potency difference criteria is required. Two-dimensional activity cliffs (2D-cliffs) have mostly been defined on the basis of Tanimoto similarity6 comparing molecular fingerprint representations2. More recently, 2D-cliffs have also been defined on the basis of substructure relationships, preferably employing the matched molecular pair (MMP) formalism7,8, leading to the introduction of MMP-cliffs9. An MMP is defined as a pair of compounds that are only distinguished by a structural change at a single site7, i.e., the exchange of a substructure, termed a chemical transformation8. For the definition of MMP-cliffs, transformation size restrictions have been introduced to limit transformations to small chemical changes typically observed in analog series9. Applying well-defined similarity and potency difference criteria, 2D-cliffs can be systematically extracted from compound databases10.
The vast majority of 2D-cliffs (i.e., close to or more than 95%, depending on the molecular representations and similarity measure used) are not formed in isolation (i.e., in the absence of structural neighbors with significant potency variations), but rather in a coordinated manner involving series of compounds with varying potency forming multiple and overlapping cliffs4,5,11. In activity cliff network representations where nodes represent compounds and edges activity cliffs, coordinated cliffs emerge as individual clusters of varying composition and size11, which can be isolated for further analysis.
In addition to 2D-cliffs, three-dimensional activity cliffs (3D-cliffs) can also be defined by comparing compound binding modes in X-ray structures12. This requires the superposition of structures of a given target available in different crystallographic ligand-target complexes and the assessment of the 3D similarity of bound ligands12. Three-dimensional activity cliffs can be further extended by taking 2D ligand information into account. This can be accomplished by systematically searching compound activity classes for analogs of 3D-cliff partners13. For example, for each cliff partner, MMPs with database compounds sharing the same activity can be determined and qualifying analogs can be assigned to the 3D-cliff13, leading to what we term herein a 3D-cliff-MMP extension. Figure 1 shows an exemplary 2D-cliff (MMP-cliff), activity cliff cluster, 3D-cliff, and 3D-cliff-MMP extension.
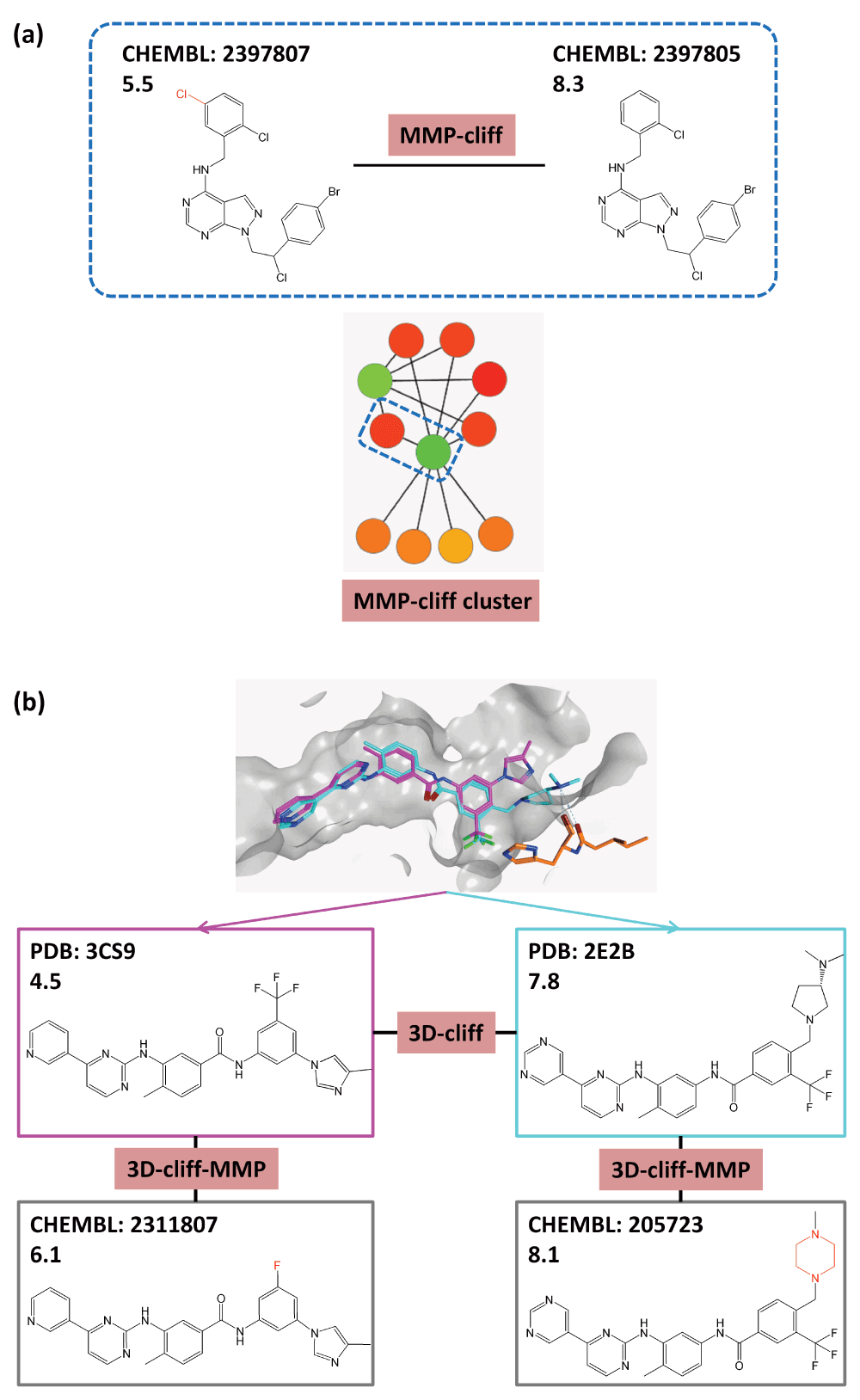
Different categories of activity cliffs are shown formed by inhibitors of tyrosine kinase ABL. MMP-cliffs are used to represent 2D-cliffs. For each compound, the ChEMBL or Protein Data Bank (PDB) ID and its negative logarithmic potency value are reported. (a) An exemplary MMP-cliff (structural modification highlighted in red) taken from an activity cliff cluster (dashed blue box) is shown. In an activity cliff network, nodes represent compounds and edges cliffs. Nodes are colored according to potency values using a continuous color spectrum from red (lowest potency) via yellow (intermediate) to green (highest potency). In network representations, coordinated activity cliffs emerge as clusters. (b) An exemplary 3D-cliff and its 2D extension are shown (3D-cliff-MMP). The extension results from MMP-based mapping of analogs from ChEMBL to 3D-cliff compounds. Structural differences between 3D-cliff compounds and their 2D (MMP) partners are highlighted in red.
In medicinal chemistry, 2D-cliffs are often considered in the context of structure-activity relationship (SAR) analysis and compound design2,3. For SAR exploration, activity cliff clusters are of particular interest because they provide more SAR information than 2D-cliffs studied individually. Furthermore, 3D-cliffs are of prime interest for structure-based design and also for computational chemistry applications including, for example, the calibration of scoring functions or free energy (perturbation) calculations. Last but not least, 2D extensions of 3D-cliffs bridge different applications in medicinal and computational chemistry and help to identify candidate compounds for further analysis.
The activity cliff information provided herein is the result of recent surveys and systematic analyses of 2D-cliffs including clusters14, 3D-cliffs15, and extensions of 3D-cliffs16. Table 1 summarizes the different activity cliff categories. All 2D-cliffs reported herein originated from the most recent release of ChEMBL (version 20)17,18 and all 3D-cliffs from the Protein Data Bank (PDB; accessed December, 2014)19. MMP extensions of 3D-cliffs were identified in ChEMBL (version 19).
Reported are the number of 2D-cliffs belonging to three different categories (FP, fingerprint; Tc, Tanimoto coefficient), corresponding activity cliff clusters (comprising at least three compounds), 3D-cliffs for different potency measurement-dependent data sets, and corresponding 3D-cliff-MMPs (giving the total number of MMPs detected for 3D-cliffs from each data set).
For all activity cliffs, an at least 100-fold difference in potency between cliff partners was consistently required. For 2D-cliffs, only (assay-independent) Ki values were considered as potency measurements. For 3D-cliffs, Ki and IC50 measurements were separately considered (using a Ki and IC50 value-based data set, respectively). In addition, 3D-cliffs were also determined in a combined Ki/IC50 data set (taking into consideration that 3D-cliffs provide a much smaller knowledge base than 2D-cliffs; vide infra). For 3D-cliff analysis, a crystallographic resolution limit of 3.0 Å was applied.
Two-dimensional activity cliffs were determined using three different molecular representations including the extended connectivity fingerprint with bond diameter 4 (ECFP4)20, molecular access system (MACCS) structural keys21, and transformation size-restricted MMPs (MMP-cliffs)9. As similarity criteria for ECFP4- and MACCS-based activity cliffs, Tanimoto coefficient threshold values of 0.55 and 0.85 were applied, respectively2,14. For an MMP-cliff, our preferred 2D-cliff definition3,4, the formation of a transformation size-restricted MMP served as a similarity criterion. By definition 2D-cliffs do not contain stereochemical information.
For the identification of 3D-cliffs, the normalized overlap of atomic property density functions calculated for a pair of bound ligands was used as a measure of 3D similarity, taking conformational, positional, and atomic property differences into account12. An at least 80% calculated 3D similarity was required as a threshold for 3D-cliff formation12,15.
Activity cliff statistics are reported in Table 1. A total of 17,111 MMP-cliffs, 31,975 ECFP4-, and 34,813 MACCS-based 2D-cliffs were identified formed by compounds active against more than 300 targets in each case. The corresponding number of activity clusters (comprising at least 3 compounds) was 1267, 1462, and 1402, respectively. Therefore, a very large knowledge base of well-defined 2D-cliffs is currently available. In addition, on the basis of Ki and IC50 measurements, 236 and 292 3D-cliffs were detected and were formed by crystallographic ligands of 26 and 43 targets, respectively. The combined Ki/IC50 data set yielded 595 3D-cliffs for 58 targets. Although many more 2D- than 3D-cliffs are currently available, as one would expect, the number of 3D-cliffs is larger than we anticipated, hence providing substantial opportunities for structural and computational studies. Table 2 provides details for the 61 different targets for which 3D-cliffs were detected. Furthermore, more than 1000 3D-cliff-MMPs were identified for each of the Ki and IC50 data sets and 2608 for the combined set (Table 1). Hence, for many 3D-cliffs, active analogs are available whose SAR characteristics and possible interaction patterns can be explored based upon 3D-cliff information, for example, by superposing them onto 3D-cliff compounds with which they form transformation size-restricted MMPs.
A total of 61 targets are listed for which 3D-cliffs were detected. For each target, the ChEMBL ID, UniProt accession ID (UniProtID)22, and the number of available 3D-cliffs are reported. 3D-cliffs were separately determined on the basis of only Ki or IC50 measurements (available for active compounds) as well as for the combined Ki and IC50 data set (Ki/IC50). In addition, the number of MMP-cliffs (if available; defined on the basis of Ki values) is also reported for each target.
The activity cliff information described above is made freely available in four separate data files containing 2D-cliffs, 3D-cliffs, 3D-cliff-MMP extensions, and superpositions of complex X-ray structures and 3D ligands for selected targets:
(1) 2D-Cliffs_and_Cliff-Clusters.xlsx (Excel format): 2D-cliffs and clusters belonging to different categories are separately recorded using ChEMBL IDs.
(2) 3D-Cliffs.xlsx (Excel): 3D-cliffs from the Ki, IC50, and Ki/IC50 data sets are separately provided using PDB IDs for compounds and UniProt22 IDs for targets.
(3) 3D-Cliff_Extension.xlsx (Excel): Analogs of 3D-cliff compounds identified by MMP search are reported. For each of the three data sets, all 3D-cliff-MMPs are listed.
(4) Superpositions.zip (all files in MOL2 format): For each target in Table 2, superpositions of complex X-ray structures and 3D ligands are provided.
These data sets are contained in an open access ZENODO deposition23. The deposition also contains a README document that details the data organization and information provided.
In this study, we have discussed different categories of activity cliffs (including cliff extensions) and reported the distribution of cliffs belonging to these categories. Given the cliff definitions applied herein, the activity cliff information we provide as an open access deposition is up-to-date and comprehensive. We hope that this large knowledge base of activity cliffs will be helpful in the practice of medicinal chemistry and structure-based drug design as well as in further evaluating and advancing computational methods.
ZENODO: Knowledge base of two- and three-dimensional activity cliffs, doi: 10.5281/zenodo.1849023
JB designed the study, NF, YH, and DS collected, organized, and deposited the data, JB wrote the manuscript, all authors examined the manuscript.
N.F. was supported by a fellowship from the Jürgen Manchot Foundation, Düsseldorf, Germany.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 25 Jun 15 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)