Keywords
Core collection, Fast Fourier Transform, Genetic resource management, Rice, Foxtail millet
This article is included in the Japan Institutional Gateway gateway.
Core collection, Fast Fourier Transform, Genetic resource management, Rice, Foxtail millet
One of the most promising techniques for conserving the diversity of genetic resources is ex situ genebank germoplasm collection. A significant effort has been made on a global scale to preserve, characterize, distribute and utilise genetic resources in order to understand their biological phenomena and to confront the vulnerable situation regarding the sustainability of future human development1,2. As the size of germoplasm collections increase, it becomes difficult to appropriately manage and extensively evaluate them3; thus, the core collection (CC) concept4 has become a fundamental genetic resource management approach and exploits the potential of a complete collection in terms of viable data management and monetary expenses5–8.
Different CCs have different purposes, characteristics and evaluation criteria7,9–11; thus, several different algorithms and informatics tools have been developed and implemented12–15 with different approaches for satisfying particular needs of each CC. Because these CCs are constructed mainly on the basis of genotypic, phenotypic, passport or geographic data (either by individual datasets or by consensus)16, there is a lack of all-inclusive datasets; this limits the possibility of generating a CC that may satisfy most basic and applied genetic resource research programs. To the best of our knowledge, no single comprehensive dataset has been properly explored to date. One possible method to create a comprehensive dataset is to represent the available data as numerical values. Several methods exist that represent genomic information into numerical values17 and agromorphological traits (ATs) into scores18. Through this mapping process, treating each data vector as a discrete signal that can, in turn, be analysed by signal processing tools (SPTs) is possible, thus providing an effective tool for a comprehensive evaluation of datasets. We present a proof-of-concept study that shows the possibility of mapping to a discrete signal any type of data available from genetic resource collections in order to take advantage of SPTs for CC selections; this possibility provides new decision-making criteria for genetic resource management and research.
Each input data must be mapped to a numerical value. This is a fundamental process of the algorithm because it enables different datasets to be analysed together, regardless of their nature. In this manner, dissimilar passport data, single nucleotide polymorphisms (SNPs), restriction fragment length polymorphisms (RFLPs), geographic information and phenotypical traits can be included in one comprehensive dataset. To consistently represent each data type, reference tables are implemented according to the nature of each particular data: genetic information (originally represented as character elements) is now represented by a numerical vector, and trait variation, simple sequence repeat (SSR) molecular markers and passport data can be represented as either binary or normalized data depending on the quantitative/qualitative nature of the data. The original data and reference tables for this study are available in supplementary material 1. Data transformation for this study rendered a matrix containing the representation of MC samples (i1, i2, i3, …in) with (j1, j2, j3, … jm) elements each, where n is the total number of samples, and m is the number of included samples characteristics, represented by a numerical value as data(i, j).
Numerical representations of each jth data element can be treated as frequency values in m data time in such a manner that each ith sample is treated as a discrete signal. The i signal corresponds to the information behaviour from each sample. This perspective will enable the implementation of SPTs such as the discrete Fourier transform and power spectrum comparison. Although SPTs can be implemented on all data available for each sample, not all data elements contain the same informativeness value to discriminate between samples. To overcome the informative difference in each j element of data, a principal component analysis (PCA) can be performed to rearrange data into a new matrix that has the high informative elements of data at the beginning and that arranges subsequent elements according to their informativeness, discarding those whose variance equals 0. This process renders two new matrices: the original characteristics mapped vectors matrix (x) and rearranged variance value matrix (X). Matrix X, therefore, contains n samples that are formed by a numerical vector with m=m-(non informative characteristics).
The main objective of Fourier transform is the decomposition of any signal into a complex histogram of frequencies. Signal function is then represented as a vectorial function whose angle and magnitude determine a sampled point in the signal19. The original Fourier model is expressed as follows:
where x is the temporal variable, ξ it the frequential variable, i is a -1 square root and e is the natural exponent. From equation 1, a derivative can be determined for any point ξ sampled in the signal.
Fourier transform can be implemented into any complex numerical series, but in a practical sense, the computational cost increases exponentially. Thus, fast Fourier transform (FFT) is more often implemented and can be defined according to Cooley-Tukey algorithm20 as follows:
where N is the vector length, x is the temporal variable, i is a -1 square root and e is the natural exponent; in such matter that an euclidean representation - with the angle, magnitude and phase that corresponds to their position in the signal - exists for any signal dot.
Therefore, mapping any signal into a vectorial representation that contains information from every original signal dot is possible. From this complex vector, useful data can be retrieved to establish a comparison between them that indirectly represents the original signal’s juxtaposition21.
Inspired by the genomic signal processing alignment-free distance (GAFD) model22, each signal corresponding to the PCA-mapped accessions data in a set Ŝi was converted into its frequency representation by applying discrete Fourier transform. Its power spectrum was then computed. Subsequently, the distance d(i, j) for a given pair of comprehensive data signals was calculated by obtaining the mean squared error (MSE) of their respective power spectra:
Finally, a distance matrix (DM) was created by performing a pairwise comparison of all sequences in the set.
In parallel, we constructed a point-to-point (RAW) DM on the basis of the MSE given to a pair of signals prior to the PCA analysis.
Selecting a CC by this method requires the generation of a DM for each sample of the MC; this provides the interrelations among samples and enables adequate selection. A schematic of the complete workflow is presented in Figure 1.

PCA: Principal Component Analysis; FFT: Fast Fourier Transform; CC: Core Collection.
In the past, several methodological procedures have been implemented to select K elements from an MC on the basis of information provided by its DM; among such procedures, the most frequently used one is the hierarchical clustering method11. However, the current algorithm does not rely on hierarchical clustering for CC selection, instead - similar to the least distance stepwise sampling method23 - CC elements are selected by an iterative process, where r samples are selected by different criteria (which may be individually implemented) on each iteration.
Selection criteria (based on the DM without hierarchical clustering) for the current algorithm is as follows:
• a) The ith sample with the most lower distance values among jth elements.
• b) The ith sample with the most higher distance values among jth elements.
• c) The ith sample with a lower distance average.
• d) The ith sample with a higher distance average.
• e) The ith sample with a lower overall distance.
• f) The ith sample with a higher overall distance.
In cases where multiple samples share selection values, an appearance priority will complete the criteria.
An example of selection process is presented in Figure 2 and its final result is presented in Figure 3.

First three principal component’s distribution of Rdata (a), methodology’s first (b), second (c) and third (d) iterations; final K=72 distribution is presented in (e).
Once the selected samples (r) are included in the future CC, they (along with others that are identical to them (s)) are removed from X for the next iteration; then, a DM2 with n2 = n–r–s is calculated. This process will continue Z times until R >= K, where R = (r1 + r2….rZ ) and K = predefined CC elements desired.
As discussed previously, the best way to evaluate a CC depends on the purpose of that CC, and even if it can be evaluated from the same dataset from which it was constructed, evaluating it with a different dataset7 is desirable. In this study, we use other datasets for our evaluation whenever possible. The list given below provides the evaluation parameters implemented in this study.
a) The average distance between each MC sample and the nearest CC sample (ANE) can be calculated using the equation as follows:
where K is all CC elements, k is each CC element and D is the distance between k and each jth cMC element whose closest CC element is k, including itself, thus rendering L total comparisons. The ideal ANE value is 0, where each sample of the CC represents itself and those similar to it. This parameter evaluates the homogeneity of the represented MC diversity.
b) The average distance between each CC sample and the nearest CC sample (ENE) can be calculated using the equation as follows:
where K is all CC elements, k is each CC element and D is the distance between k and its closest CC element cCC, excluding itself, in L total comparisons. With such an evaluation parameter, higher dispersion renders higher scores with the aim of evaluating the dispersion among selected CC elements.
c) The average distance between CC samples (E) can be calculated using the equation as follows:
where K is all CC elements, k is each CC element and D is the distance between k and all other jth CC elements cCC, excluding itself, in L total comparisons. This evaluation parameter indicates higher scores when CC elements have greater distances between themselves.
While previous evaluation parameters are useful for data dispersion analysis, such parameters will not evaluate how well the distribution of the MC is represented on the CC; therefore, the distribution comparisons tests that were included are as follows:
d) The homogeneity test (F – test for variances and t – test for means; α = 0.05) between the CC and MC for each trait can be represented as a percentage of traits that are statistically different (MD for means and VT for variances)9.
e) The coincidence rate (CR) can be calculated using the equation as follows:
where R is the range of each m trait, and M represents the number of traits.
f) The variable rate (CV) can be calculated using the equation as follows:
where CV is the coefficient of the variation of each m trait in the CC and MC, and M is the number of traits.
According to Hu et al.10, a valid CC has CR > 80 and MD < 20, which are the limits for the ideal representation of the identity and distribution of the MC.
g) The alleles coverage (CA) can be calculated using the equation as follows:
where ACC is a set of alleles in the CC, and AMC is a set of alleles in the MC; ACC measures the percentage of alleles from the MC that are present in the CC12.
To compare the obtained CCs with an established methodology, we implemented Core Hunter 2 (CH)13 as a reference and used it with the program’s default parameters on the agrological and genomic datasets.
To determine the efficiency of the analysis of data behaviour by a point-to-point direct comparison, a synthetic dataset was constructed using binary data (Sdata) with manageable n and m elements (supplementary material 1).
To test the algorithm in real biological-context scenarios, the CCs from different MCs were constructed and evaluated.
To test the algorithm’s CCs versus the scores of the MCs, 780 rice (Oriza sativa(L.)) accession and 423 foxtail millet (Setaria italica subsp. italica (L.) P. Beauv.) accession data were retrieved from the National Institute of Agrobiological Sciences (NIAS) http://www.gene.affrc.go.jp/databases_en.php.
According to the available data, different datasets were assembled. The 762 SNPs from the 780 rice accessions retrieved from the NIAS database (Rdata) were divided arbitrarily into two subsets of 331 SNPs each for constructing two smaller datasets (RdataI and RdataIII). In addition, ATs were categorized and mapped into the binary data for 273 of the 780 accessions, resulting in 38 variables (RdataII). The variables from the 423 foxtail millet genotypes with transposon displays24 were used as a single dataset (Fdata). For a subset of 141 accessions (FdataI), 9 ATs were categorized and mapped into binary data, resulting in 28 variables (FdataII). The substitution tables used during this mapping are presented as supplementary material 1.
All methodological procedures (except for CH, which was implemented according to the software’s default parameters and which is available for download at www.corehunter.org) were performed using FreeMat v4.2 (www.freemat.sourceforge.net). All original codes are available as supplementary material 1.
The selection criteria were chosen to look for the best possible distribution of selected CC elements within the DM. Although hierarchical clustering has proven to be an effective method for determining collection structure and sampling CC25 and although it has been implemented in different crops26,27 and included in various selection algorithms11, hierarchical reconstruction presents the challenge of selecting an appropriate model for biological interpretation that can be applied to everything from unweighted pair-group averages to Markov models in Bayesian estimations28. To avoid the challenge of selecting a reconstruction model, we decided to work strictly with the DM. By selecting the items described in this methodology, we aimed to retrieve representative elements from among the distributions of collections; however, because of its iterative nature, this methodology may render high redundancy under certain data distributions. Despite this limitation, the methodology has proven to be capable of selecting representative elements of the MC’s diversity.
Evaluation criteria were applied according to Odong et al.7 without excluding the classic criteria used in 9,10. The selected CCs render proper results in general terms. As expected, selected CCs did not always reach for optimal values for MD and CR, this is due the fact that it is not the aim of the selection method to render a CC with similar distribution to that of the MC, but to make sure to include as much diversity as possible.
It is our belief that scoring the CC sets obtained with these methodologies will enable genetic resource banks to provide clear descriptors of what their CC strengths and limitations are with respect to the MC from which they come and will provide adequate tools for determining the possible purposes of the selected CCs.
Although several representations of genotypic characteristics (particularly those involving DNA sequences29–31) have been proposed, real-number-based mappings have not been discarded; indeed, this type of mapping has been highly studied for signal analysis even when they share two principal problems: the preferential magnitude of some nucleotides and the non-equidistance of all nucleotides32,33. The arbitrary values selected for SNPs’s numerical representation of genotypes aim to maintain equidistant relations among purines and among pyrimidines in such a manner that the same distance is also preserved between at least one of them and the undetermined values. ATs are represented as binary data. This representation may prove useful for discrete data but requires a clustering procedure for continuous data. In this study, we arbitrarily generated clusters for the latter and then represented them as the former. Although this implementation may not be the most accurate regarding biological or agronomical significance, it serves as the first approach for testing the feasibility of the use of signal processing techniques when merging several datasets to construct one CC.
The RAW comparison establishes a distance value on the basis of the average distance between each mapped value on each element while the FFT power spectra implementation compares the signals in the frequency domain. Using FFT, establishing a DM on the basis of how data ’shifted’ rather than on the basis of average point-to-point comparisons was possible. The FFT approach provides a different DM, where its compared elements are clustered based on the similarity of the shift among data, regardless of whether the shift is in the opposite phase. We believe that this procedure may reveal additional information about the relations not only between elements but also between the individual components within each element.
FFT comparisons of signals without PCA are a good approach for CC selection. Nevertheless, PCA implementation enables us to avoid possible misleads in random data arrangements, as, for example, palindromic data that could result in the same power spectra. Moreover, through PCA, we could organize data according to their levels of impact on the difference between accessions, which - when their magnitudes were obtained - inherently rendered a representation of informativity relations among values. This ’data behaviour’ was used as the element for pairwise comparisons, and although this approach clusters differently from RAW comparisons, we believe that it will provide a new perspective for CC selection and open the possibility of further data exploration.
Our first approach was to measure the comparisons under different K values. We compared the approach of the RAW signals with the PCA-FFT-treated signals. Results from Sdata, Fdata and Rdata are presented in Table 1–Table 3. As expected, most evaluation criteria improved as K increased.
The use of FFT signals renders better overall scores than the use of RAW signals in Sdata and Fdata; however, this advantage diminishes in Rdata. We speculate that this difference can be explained by the mapping procedures used; further research regarding this matter is encouraged.
Using the CHs’ rendered K values, we used both CH and FFT to generate the CCs on the above datasets. The evaluation of these CCs is summarized in Table 4 and in Figure 4, Figure 5. Both methodologies rendered similar results, yet PCA rendered better results on parameters representing MC distribution; this could be an effect of the selection method’s intrinsic redundancy.

First two principal component’s distributions of k=11 CC (orange) selected by CH (a), PCA (b) and RAW (c) in Sdata distribution (blue).

First two principal component’s distributions of k=84 CC (orange) selected by CH (a) and PCA (b) in Fdata distribution (blue).
Thus far, the proposed CC selection method and algorithm appear worthy of further exploration. We are aware that two particular fundamental elements require immediate attention. First, a better mapping solution for both genotypic and AT numerical representation needs to be determined. Second, the selection system developed by us is directly based on the DM and is prone to high redundancy in some data distributions. As discussed earlier, this selection system was chosen in order to avoid the problems associated with hierarchical clustering and further allocation selections13,34. Both issues should be addressed in the near future.
To demonstrate that FFT-based CC selection can include and analyse data regardless of its origin, we concatenated corresponding signals from FdataI with FdataII as well as RdataI and RdataIII with RdataII to construct MFdata, MRdataI and MRdataIII. The comprehensive sets were used to construct CCs; the sets were then compared with both their original genotype and phenotype MCs. These comparisons are shown in Table 5–Table 8, and their distributions are represented in Figure 6–Figure 9.
| vs RdataII | |||
|---|---|---|---|
| RdataII | MRdataI | MRdataIII | |
| K | 24 | ||
| ANE | 0.4594 | 0.4652 | 0.4618 |
| ENE | 0.4796 | 0.4896 | 0.4742 |
| E | 0.6402 | 0.6205 | 0.6169 |
| MD | 0 | 5.2632 | 0 |
| VT | 39.4737 | 42.1053 | 60.5263 |
| CR | 63.8082 | 61.8988 | 68.2437 |
| CV | 3.8262 | 2.2285 | 4.1332 |
| AR | 95.4268 | 98.7805 | 98.7805 |
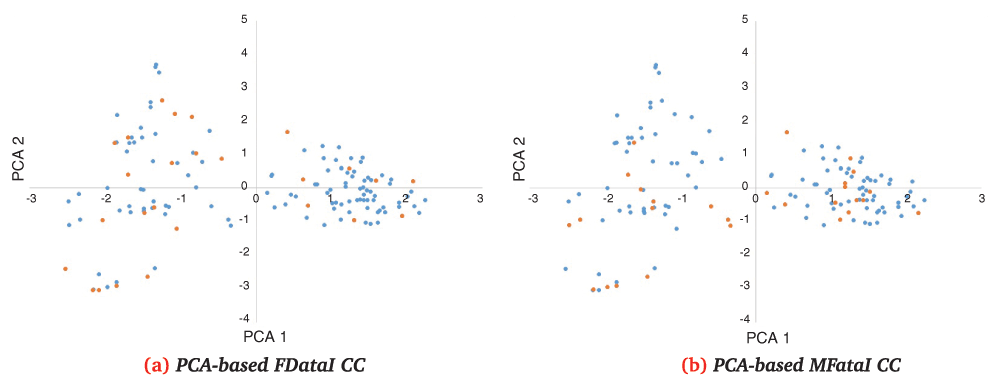
First two principal component’s distributions of k=24 CC (orange) selected by PCA from FDataI (a) and MDataI (b) in FdataI distribution (blue).

First two principal component’s distributions of k=24 CC (orange) selected by PCA from FDataII (a) and MDataI (b) in FdataII distribution (blue).
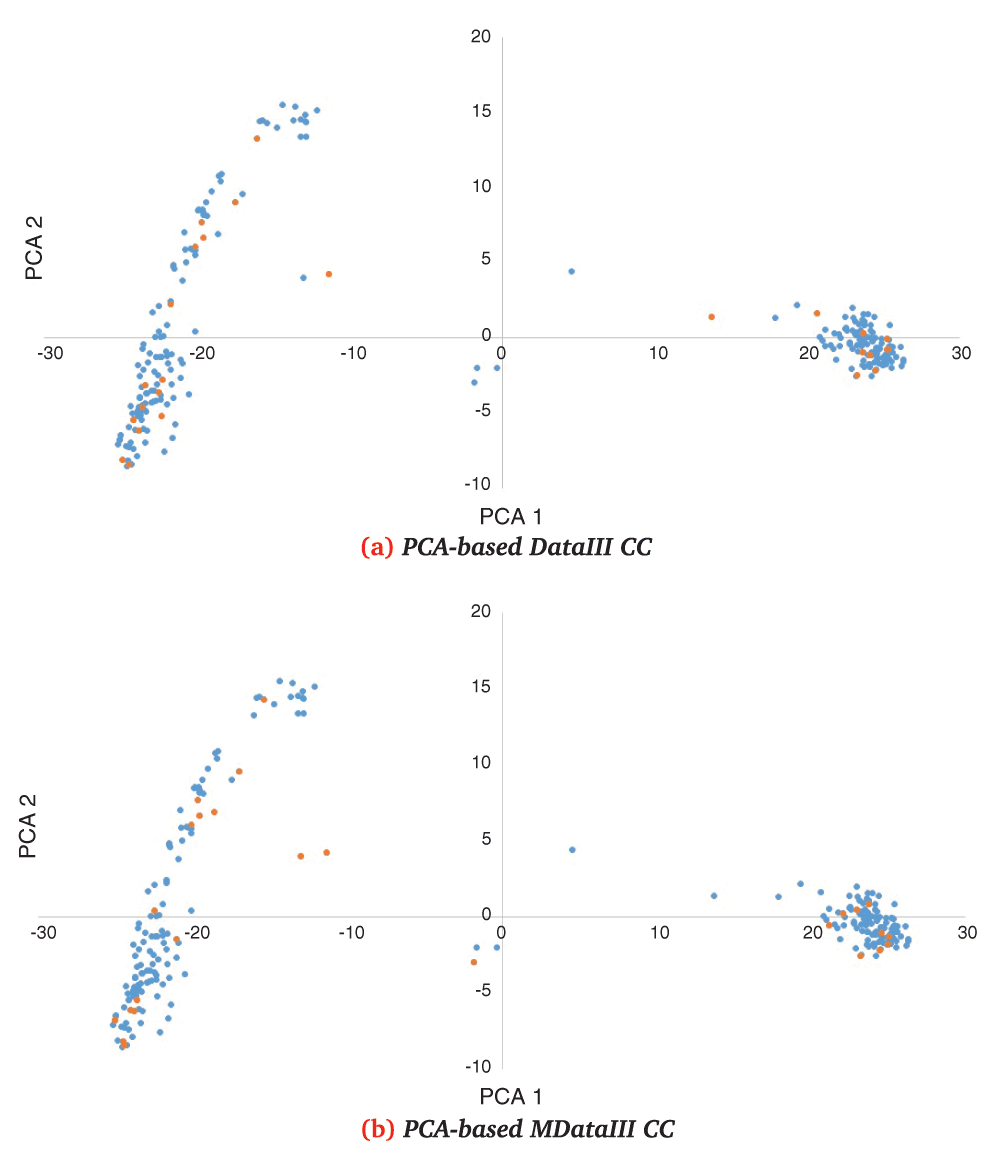
First two principal component’s distributions of k=24 CC (orange) selected by PCA from RDataIII (a) and MDataIII (b) in RDataI distribution (blue).

First two principal component’s distributions of k=24 CC (orange) selected by PCA from RDataII (a) and MRDataI (b) in RdataII distribution (blue).
These comprehensive CCs showed overall better scores than genotypic-only CCs when compared with genotypic-only data. On the contrary, there was a better overall score in phenotypic-only CCs when compared against phenotypic-only data. In the latter case, it should be kept in mind that comprehensive data also consider genotypic data; this could explain why better selections are made when only phenotypic data are considered because genotypic variations may reduce the impact of some phenotypic traits in the PCA analysis.
The generation of a DM based on signal comparisons originating from mixed data construction enables us to explore one of the most interesting applications of this algorithm. By mapping genotypic and AT data, constructing a single signal with all data available for a particular accession is possible. The possibility of including genotypic data with phenotypic traits, geographical locations, climates, habitats, nutritional requirements, symbiotic relationships and so forth provides an opportunity for determining the best information to be included in the selection process in order to cope with the particular objectives for which that CC is being selected. This concept, in addition to adequate scoring systems, may prove useful in designing tailored CCs that comply with specific research/breeding objectives.
The use of SPTs in CC selection, as presented in this algorithm, enables us to analyse all available data comprehensively and from different perspectives. Despite its limitations, this signal construction makes it possible to analyse all available data regarding each accession in CC selection with good results. The efficiency of SPTs in CC selection suggests that the use of these tools in MC analysis may provide useful information not only for CC but also for other purposes. The implementation of current and other SPTs in all-inclusive MC-mapped signals is worth further exploration, and we believe that it will be an important asset to genetic resource management and exploitation.
EB conceived and designed the algorithm, performed the implementation, analysed the data and wrote the manuscript. MT contributed to the design of the algorithm, data analysis and manuscript drafting and correction. Both authors have read and approved the final manuscript.
This research is supported in part by the SATREPS project by JST and JICA, Diversity Assessment and Development of Sustainable Use of Mexican Genetic Resources and in part by JSPS Grant-in-Aid 25257416.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We would like to thank F1000Research for the free-submission prize awarded at ISCB-ASIA 2014.
Genomic and AT numerical mapping equivalences and Original Codes
SyntheticData.cvs : Synthetic Data representing binary information.
FoxtailATMap.cvs : Mapped Rice AT where 1=’presence’; 0=’absence’ (corresponds to FdataII).
FoxtailGenMap.cvs : Mapped Foxtail Genotype where 1=’presence’; 0=’absence’ (corresponds to FdataI).
RiceATMap.cvs : Mapped Rice AT where 1=’presence’; 0=’absence’ (corresponds to RdataII).
RiceGenIMap.cvs : Mapped RICE SNPs where 1="T"; 2="C"; 4="N"; 6="G" and 7="A" (corresponds to RdataI).
RiceGenIIIMap.cvs : Mapped RICE SNPs where 1="T"; 2="C"; 4="N"; 6="G" and 7="A" (corresponds to RdataIII).
Codes are enclosed as scripts in FFTCoreCollCodes.rar, available online at https://zenodo.org/badge/doi/10.5281/zenodo.16895.svg Refer to README.m for details.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
At the request of the author(s), this article is no longer under peer review.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)