Keywords
auxin, cell division, gene
auxin, cell division, gene
The plant hormone auxin plays a vital role in nearly every aspect of plant growth and development1. Auxin-responsive gene expression relies on the TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX (TIR1/AFB) pathway to trigger the expression of genes controlling auxin-regulated cell division, expansion, and differentiation2,3. Whereas the role of the TIR1/AFB pathway in the auxin signal transduction pathway has been well established, the existence of additional components raises the possibility that we have yet to uncover the entire story of auxin signaling.
Auxin signaling through the TIR1/AFB pathway involves three major protein families (Figure 1A) – the auxin-binding TIR1/AFB F-box proteins, the AUXIN RESPONSE FACTOR (ARF) transcription factors, and the AUXIN/INDOLE-3-ACETIC ACID INDUCIBLE (Aux/IAA) repressor proteins2,3. In the absence of auxin, the Aux/IAA proteins repress activity of the ARF transcription factors4. In the presence of auxin, the TIR1/AFB F-box proteins, which participate in a SCF (Skp1-Cullin-F-box) E3 ubiquitin ligase, interact with Aux/IAA repressor proteins to form a co-receptor, with auxin acting as the “molecular glue”4–6. This interaction results in ubiquitylation and consequent degradation of the Aux/IAA repressor proteins through the 26S proteasome, relieving repression of the ARF transcription factors and allowing for auxin-regulated gene transcription7. Interactions among these three protein families is now understood at a molecular level2 and provides a signal transduction pathway that controls auxin-responsive gene transcription in plants. For recent reviews of the TIR1/AFB pathway, please see 1,3.
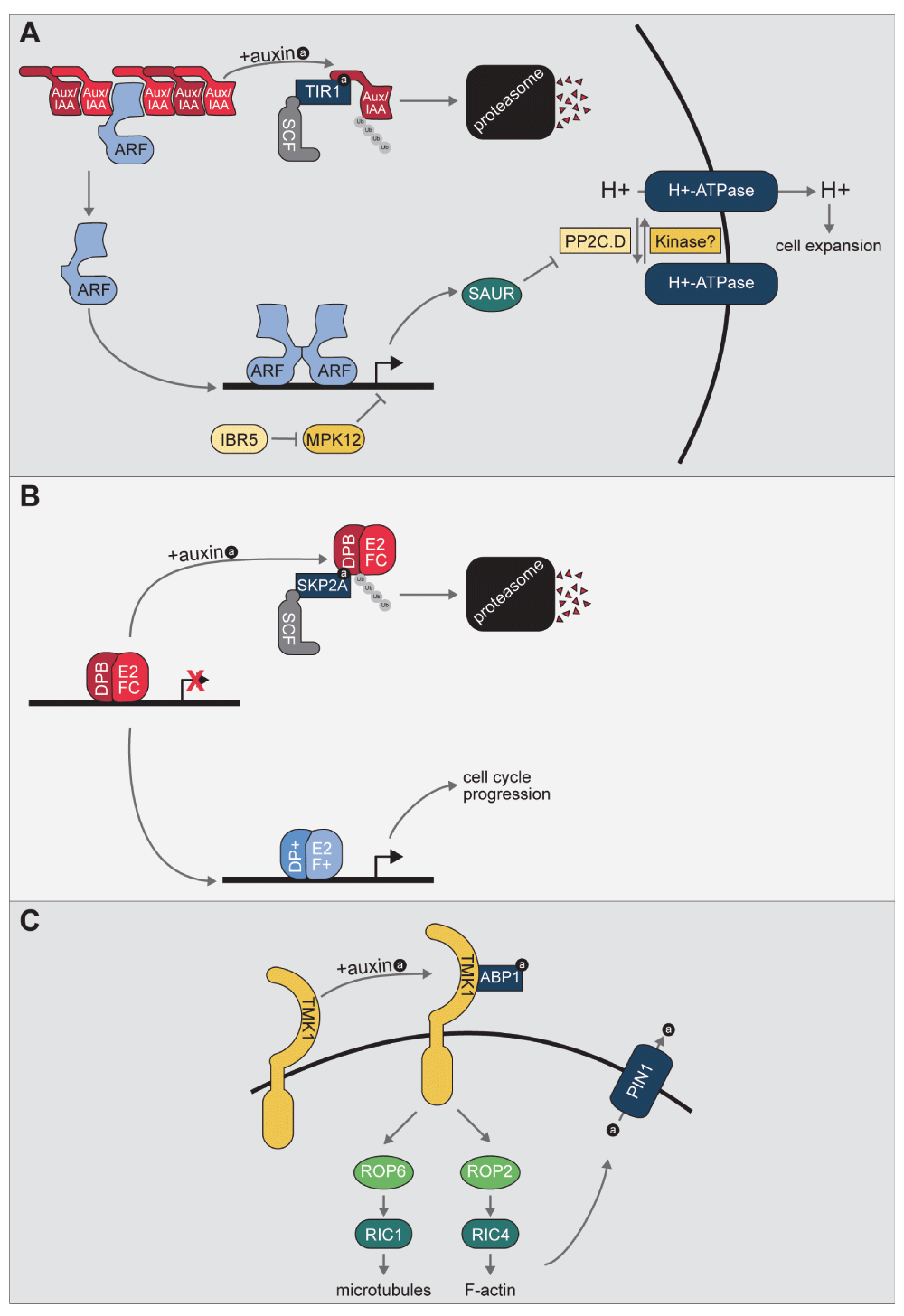
(A) Model of the TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX (TIR1/AFB) signaling pathway. Auxin promotes the formation of the TIR1/AFB Auxin/INDOLE-3-ACETIC ACID INDUCIBLE (Aux/IAA) co-receptor to promote the ubiquitylation and subsequent degradation of the Aux/IAA repressor. Aux/IAA degradation relieves repression of AUXIN RESPONSE FACTOR (ARF) transcription factors, allowing for auxin-responsive gene expression. One of the transcript families upregulated by auxin is the SAUR family. The small SMALL AUXIN UP RNA (SAUR) proteins encoded by these transcripts have been suggested to play roles in multiple processes, one of which is interaction with and inhibition of members of the PP2C.D family of phosphatases, which act to regulate H+-ATPase activity. Further, INDOLE-3-BUTYRIC ACID RESPONSE5 (IBR5) and MITOGEN-ACTIVATED PROTEIN KINASE12 (MPK12) have been implicated in regulating auxin-responsive gene transcription; this regulation is not through destabilization of the Aux/IAA repressors, suggesting a yet-to-be discovered mechanism of regulating auxin-responsive gene expression. For in-depth reviews of the TIR/AFB signaling pathway, please refer to 1,3. For an in-depth review of SAUR proteins, please refer to 15.
(B) Model of the S-PHASE KINASE ASSOCIATED PROTEIN 2A (SKP2A) signaling pathway. E2FC/DPB repress expression of cell cycle genes. The F-box protein SKP2A binds auxin and promotes degradation of E2FC/DPB in an auxin-dependent manner. Degradation of E2FC/DPB relieves repression of cell cycle genes and allows for binding by activating E2F+/DP+ complexes. For an in-depth review of the SKP2A signaling pathway, please refer to 8.
(C) Model of the putative AUXIN BINDING PROTEIN1 (ABP1) signaling pathway. Apoplastic auxin is bound by ABP1, which allows for interaction with the TRANSMEMBRANE KINASE RECEPTOR (TMK) family of leucine-rich repeat receptor-like kinases. ABP1 binding of auxin regulates RHO-LIKE GTPASE 2 (ROP2) and ROP6 activation and binding to ROP interactive CRIB motif-containing proteins (RICs) proteins to positively regulate microtubule polymerization and F-actin polymerization and also alter PINFORMED1 protein localization to alter auxin efflux. For an in-depth review of the ROP/RIC system, please refer to 54.
In addition to components of the TIR1/AFB pathway, other factors implicated in auxin response have been identified – however, roles for these components in regulating auxin signaling are much less understood. Although insight into the function of some of these factors, such as S-PHASE KINASE-ASSOCIATED PROTEIN 2A (SKP2A), SMALL AUXIN UP RNAs (SAURs), INDOLE 3-BUTYRIC ACID RESPONSE5 (IBR5), and AUXIN BINDING PROTEIN1 (ABP1), has been elusive, roles for these factors in regulating auxin outputs are slowly starting to be determined. In this commentary, we explore recent advances in our understanding of these factors and their roles in auxin response.
Whereas extensive work has gone into understanding the auxin-binding capabilities of TIR1 and its downstream effects on auxin-regulated developmental responses (Figure 1A), less is known about the molecular mechanisms that directly connect auxin to its role in cell division. Several cell cycle genes are upregulated by auxin8 and other cell cycle genes contain auxin response elements in their promoter regions; however, these do not appear to be upregulated by auxin8, suggesting that additional mechanisms linking auxin to cell division control may exist. Heterologously expressed SKP2A, an F-box protein involved in regulating the proteolysis of cell-cycle-related transcription factors9,10, directly binds auxin11 and may provide a mechanism for auxin-mediated regulation of cell division (Figure 1B).
The retinoblastoma-E2F pathway regulates the cell cycle through the interaction of E2F proteins with dimerization proteins (DPs) to form transcription factors that either activate or repress the expression of genes involved in cell cycle progression12. SKP2A incorporates into an SCF complex9,13 with E3 ubiquitin ligase activity13. Intriguingly, binding of auxin by SKP2A is necessary for degradation of E2FC and its dimerization partner DPB through the 26S proteasome11. Degradation of E2FC and DPB relieves repression of cell cycle control genes to allow cell cycle progression9,10. In addition to regulating the degradation of E2FC/DPB, auxin binding promotes proteolysis of SKP2A itself13, perhaps setting up a system in which auxin prevents SKP2A overfunction11. The skp2a mutant hyperaccumulates E2FC and DPB protein10,13. Further, expressing a SKP2A variant unable to bind auxin in the mutant background fails to rescue this phenotype, suggesting that auxin binding by SKP2A is required for E2FC/DPB degradation11. Overexpression of SKP2A results in increased cell division and induces lateral root primordia (LRP) formation, a process known to be dependent on auxin signaling9. The molecular phenotypes of the skp2a mutant combined with the phenotypes of the SKP2A overexpression lines suggest a role for SKP2A in promoting auxin-regulated cell division.
Involvement of SKP2A in auxin binding and consequent degradation of the cell-cycle regulators E2FC and DPB implicate this F-box as a missing link connecting auxin regulation to cell division; however, relatively little is known about this pathway. Many questions remain for SKP2A roles in auxin-regulated cell division control. For example, SKP2A directly binds auxin and functions as part of an SCF complex responsible for targeting downstream components for degradation – are additional factors, other than E2FC and DPB, targeted for degradation by SKP2A to contribute to auxin response? Further, are activating E2F+/EP+ complexes similarly regulated by the proteasome? What is the effect on plant growth and development if this auxin-induced degradation of these factors is disrupted? There appear to be no gross effects on plant morphology in the skp2a mutant; is this because of redundancy or is it indicative of a minor role for SKP2A in plant growth and development? Answering these questions will inform our understanding of SKP2A roles in mediating auxin effects on cell division.
Auxin regulation downstream of the TIR1/AFB pathway involves the induction of the early auxin response gene family SAURs. Initially discovered as auxin-induced transcripts in elongating soybean hypocotyls using a hybridization screen14, multiple lines of evidence have been used to assign SAUR functions in auxin-related aspects of plant growth and development, including cell expansion, tropic growth, and apical hook development15. Although SAURs represent the largest family of early auxin response genes, SAUR function in mediating auxin effects has only recently begun to be elucidated.
Ca2+ is a well-known secondary messenger regulating developmental and physiological aspects of plant growth16. Auxin has been proposed as a Ca2+ activating signal17; however, mechanisms connecting Ca2+ to auxin signaling have remained largely elusive. SAUR proteins from multiple species interact with calmodulin (CaM) in a calcium-dependent manner18–20 and additional SAUR proteins are predicted to contain a CaM-binding site15. Binding of SAUR70 to CaM or CaM-like proteins has been confirmed in planta19; however, further studies will be necessary to determine the extent and functional relevance of SAUR-CaM interactions in auxin-regulated calcium signaling.
Several SAUR subfamilies, including SAUR19-24, are likely involved in cell expansion21; however, mechanistic insight into SAUR roles in this auxin-regulated response has been lacking. Auxin has long been proposed to induce cell elongation through an acid growth mechanism22, in which auxin is responsible for activating H+-ATPases to acidify the extracellular matrix, in turn activating expansins and promoting solute and water uptake to drive cell expansion. Recently, Spartz et al.23 determined that SAUR proteins promote phosphorylation of the C-terminal autoinhibitory domain of PM H+-ATPases, causing activation. In addition, several SAUR proteins interact with a group of the D-clade PP2C phosphatases to inhibit phosphatase activity23,24. These phosphatases likely modulate the phosphorylation status of H+-ATPases23. Thus, SAURs likely act to inhibit the PP2C.D inhibitors of H+-ATPase activity, suggesting a role for the SAUR proteins as positive effectors in auxin-mediated cell expansion through regulation of the PM H+-ATPase activity. If PP2C.D deactivates H+-ATPase activity through de-phosphorylation, it then follows that a kinase is necessary to activate H+-ATPases. Many protein kinases have been proposed to regulate H+-ATPase activity25 – perhaps one of these functions in an auxin-dependent manner to regulate H+-ATPases. Further, the identified SAUR-CaM interactions combined with the recently identified roles for SAUR proteins in regulating phosphatases raise the possibility that SAUR proteins could act as a link between calcium signaling, auxin, and phosphatase activity.
Recent studies have provided increasing evidence of SAURs’ importance in auxin-regulated plant growth and development. The large families of these proteins identified in widespread plant species suggest multiple and diverse SAUR functions in auxin response. One possible mechanism for SAUR regulation of such varied auxin-related plant responses may be provided by combinatorial diversity in SAUR-PP2C.D interactions. In Arabidopsis, there are 81 SAURs26 and nine PP2C.D family members24, many of which have been proposed to have differential expression patterns throughout the plant27. Varying SAUR-PP2C.D combinations may regulate the phosphorylation status of distinct downstream elements to regulate different aspects of auxin response15. Interaction experiments using the different SAUR-PP2C.D combinations may uncover whether SAUR proteins from additional clades also interact with these phosphatases. Further, the distinct subcellular localization and various developmental processes associated with individual SAUR proteins15 suggest that SAUR targets in addition to H+-ATPases likely exist. Whereas recent studies have finally begun to illuminate SAUR molecular functions in auxin response, further research will surely uncover additional mechanisms connecting the SAUR proteins to auxin signaling.
Auxin pathway roles for IBR5 and its interacting MAP kinase MPK12 remain enigmatic. The ibr5 mutant was initially isolated for its resistance to the auxin precursor indole-3-butyric acid (IBA)28, but subsequent studies revealed that ibr5 was resistant to all tested auxins29 and auxin transport inhibitors30. In addition to reduced physiological responses to exogenous auxin application, ibr5 mutants display developmental phenotypes29,31,32 and reduced auxin-responsive transcription29–32, suggesting roles for IBR5 in the auxin signaling pathway.
IBR5 encodes a dual-specificity protein phosphatase29; related phosphatases de-phosphorylate MAP kinases33,34. Dual-specificity protein phosphatases are distinct from the PP2C-type phosphatases associated with SAUR activity (see above). IBR5 splice variants appear to play distinct roles in regulating plant growth and auxin responses and at least some of these roles may be independent of its catalytic activity31. However, the IBR5 catalytic cysteine is necessary for auxin responsive inhibition of root elongation30,31 and IBR5 can de-phosphorylate the Arabidopsis MPK12 in vitro35. Further, RNAi lines of MPK12 display auxin resistance35, suggesting that IBR5 and MPK12 play opposing roles in regulating auxin responsiveness.
ibr5 double mutants with other mutants that dampen auxin responses, including tir1, axr1, and aux1, exhibit additive auxin resistance in one or more bioassays30. Most notably, combining ibr5 with an auxin receptor mutant, tir1, greatly enhances auxin resistance relative to either parent30, consistent with the possibility that IBR5 effects on auxin response are TIR1 independent. Similar to other auxin-resistant mutants, ibr5 exhibits decreased levels of auxin-responsive transcripts29–32. However, unlike other characterized auxin-response mutants, Aux/IAA proteins are not stabilized in ibr5 and are actually destabilized30,31, again suggesting that IBR5 modulates auxin signaling in a manner unique from other known auxin response regulators. ARF proteins mediate auxin-responsive gene transcription and the primary known mechanism of ARF regulation is repression by Aux/IAA proteins3. The decreased auxin-responsive gene transcription29–32 combined with destabilized Aux/IAA repressor proteins30,31 observed in ibr5 mutants suggest that either Aux/IAA destabilization is not the sole mechanism of regulating ARF activity or that there exists an additional auxin signaling pathway that ends in regulating the same gene targets as the TIR1/AFB pathway. Elucidating IBR5 and MPK12 targets may help differentiate between these possibilities. Recently, IBR5 was found to interact with SUPPRESSOR OF G2 ALLELE SKP1 (SGT1b), HEAT SHOCK PROTEIN90 (HSP90), and the Toll/interleukin-1 receptor domains of CHILLING SENSITIVE 3 (CHS3), SUPPRESSOR OF NPR1-1 (SNC1), and RESISTANT TO P. SYRINGAE 4 (RPS4)36. IBR5 interaction with SGT1b may provide a mechanism for IBR5 regulation of auxin responses; mutants defective in SGT1b/ETA3 have been isolated as enhancers of tir1 auxin resistance, perhaps by modulating 26S proteasome activity37. SGT1b is a co-chaperone with HSP90. Further, TIR1 has recently been identified as a HSP90 client and HSP90 plays roles in integrating temperature and auxin signaling38; perhaps IBR5 is an additional HSP90 client to allow temperature and auxin response integration.
Opinions about ABP1, first discovered over 40 years ago, have varied over the years. ABP1 has been proposed to act as an apoplastic auxin receptor whose downstream signal transduction pathway regulates cytoskeletal rearrangement and internalization of auxin transporters in response to auxin39–41. Study of roles for ABP1 in plant auxin signaling was limited by the reported embryo lethality of null abp1 alleles42–45. In the absence of a viable allele, the field made progress in understanding ABP1 function by use of ABP1 knockdown lines, provided by expression of an inducible ABP1 antisense transcript or of an inducible single-chain fragment variable from a monoclonal antibody raised to ABP146. The identification of an EMS-generated TILLING line, abp1-5, carrying a point mutation in the auxin-binding pocket of ABP147,48 allowed for intense study of ABP1 functions in auxin signaling and spurred an explosion of ABP1-related discoveries, uncovering roles for this elusive auxin-binding protein in a wide variety of processes throughout plant development39. These new findings allowed for widespread acceptance of ABP1 as a bona fide auxin receptor41.
The recognition of ABP1 as an auxin receptor, however, has once again been called into question. A recent report showing that new abp1 null alleles display no obvious auxin-related or developmental phenotypes49 was contradictory to earlier reports of the embryo lethality of the abp1-1 null allele42–45 and the physiological and molecular phenotypes displayed by conditional knockdown and abp1-5 alleles39. This conflict was recently partially reconciled by the discovery that the embryo lethal phenotypes of abp1-1 and abp1-1s were caused by a loss of the neighboring BELAYA SMERT gene rather than from loss of ABP150,51 and that the abp1-1 allele, which is null for ABP1, displays no obvious morphological phenotypes when the BSM defect is rescued in the mutant50. Further, at least some of the phenotypes observed in the abp1-5 TILLING allele may be the result of a background mutation in PHYTOCHROME B52. Future work with new ABP1 genetic resources49,51 will be necessary to clarify the role of ABP1 in auxin signaling and plant development and will determine whether abp1 null alleles display molecular phenotypes.
The auxin-binding affinity of ABP1 combined with its widespread conservation throughout plants would suggest an important role for ABP1; however, lack of physiological phenotypes in the null mutants49,50 would suggest that ABP1 does not play a prominent role in Arabidopsis development. At this time, auxin-related roles for downstream components in the ABP1 pathway (Figure 1C) including the TRANSMEMBRANE KINASE (TMK) family of receptor-like kinases53 and the ROP-RIC system54 remain unchallenged. This is a turbulent time in the ABP1 field as new discoveries are being made and roles (or lack thereof) for this signaling pathway in plant growth and development are being clarified. Roles for ABP1 in auxin response and development have once again become controversial; clearly more work will be needed to reconcile conflicting reports in this area.
The existence of factors in addition to the components of the well-established TIR1/AFB pathway suggests that we have yet to uncover the entire story of auxin response. Recent advances in the field are slowly bringing to light roles in auxin signaling for each of these factors − SKP2A, the SAUR proteins, IBR5, and ABP1 – however, many questions still remain. In addition, although the TIR/AFB pathway appears to be well characterized, new structural data suggest that there are additional regulatory aspects of this pathway, including ARF proteins acting as molecular DNA calipers and ARF and Aux/IAA protein multimerization2,55 that have yet to be fully explored. Further research will undoubtedly uncover new regulatory mechanisms for the TIR/AFB pathway and molecular roles for these untethered factors in auxin signaling − the sky is the limit!
ABP1, AUXIN BINDING PROTEIN1; AFB, AUXIN SIGNALING F-BOX; ARF, AUXIN RESPONSE FACTOR; Aux/IAA, Auxin/INDOLE-3-ACETIC ACID INDUCIBLE; bHLH, basic Helix-Loop-Helix; CaM, Calmodulin; DP, Dimerization Protein; IBR5, INDOLE-3-BUTYRIC ACID RESPONSE5; MPK12, MITOGEN-ACTIVATED PROTEIN KINASE12; ROP, RHO-LIKE GTPASE; SAUR, SMALL AUXIN UP RNA; SCF, Skp1-Cullin-F-box; SKP2A, S-PHASE KINASE ASSOCIATED PROTEIN 2A; TIR1, TRANSPORT INHIBITOR RESPONSE1; TMK, TRANSMEMBRANE KINASE RECEPTOR.
This research was supported by the William H. Danforth Plant Science Fellowship Program (to S.K.P.), the National Science Foundation (IOS-1453750 to L.C.S.) the National Institutes of Health (R01 GM112898-01 to L.C.S.).
We are grateful to Tara Enders, Elizabeth Frick, and Marta Michniewicz-Paciorek for critical comments on the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
|
Version 1 03 Feb 16 |
read | read | read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)