Keywords
Yersinia pestis, yersiniae, ail locus, pH6 antigen, virulence factors
Introduction
Across an infection timeline, the host and invading bacterial pathogen each vie for supremacy. At any given time, either may have the upper hand, but the final outcome of this battle ultimately determines the fate of the host. The triggered host response will aim to reduce the infectivity of the pathogen, but in order to stay one step ahead many bacterial species have evolved sophisticated strategies to ensure they can successfully cause infection following colonisation.
The three human pathogens belonging to the genus Yersinia employ a range of virulence factors that confer efficient adherence to host cells/tissues and subvert host cell functions. This mini-review highlights the key virulence factors that constitute the virulence arsenal of Yersinia spp. and how such a sophisticated suite of biological weapons enables these pathogens to combat host defences.
Yersinia pseudotuberculosis, Yersinia pestis, and Yersinia enterocolitica are highly adaptable psychrotrophic primary human pathogens. Y. pseudotuberculosis and Y. enterocolitica cause self-limiting gastric infections. Y. pestis is a recently evolved near-identical subclone of Y. pseudotuberculosis1,2 with approximately 98% identity at the DNA level. Its strategy for transmission relies on the colonisation of rat fleas, which then carry Y. pestis between the rodent host and humans3. Once inside the human host, Y. pestis can cause bubonic, pneumonic, and septicaemic plague with mortality rates approaching 100% without antibiotic treatment4. The World Health Organisation considers Y. pestis a ‘re-emerging’ pathogen that, worryingly, is capable of acquiring resistance to multiple antibiotics5 and is also a serious potential bioterrorism threat. The differences in lifestyle and virulence between Y. pseudotuberculosis and Y. pestis are mostly attributable to minor genomic differences on the respective chromosomes and the presence of two additional virulence plasmids that Y. pestis possesses.
The Yersinia type three secretion system
The key Yersinia virulence determinants and certainly the most comprehensively studied are those secreted via a type three secretion system (T3SS). To evade host innate immunity and to enable the pathogen to replicate and propagate extracellularly, all human pathogenic Yersinia species harbour an approximately 70 kb virulence plasmid. Located on this plasmid is a set of genes whose transcription is activated by temperatures of 37°C in the presence of millimolar concentrations of calcium, conditions representing the mammalian host. These genes code for the T3SS ‘nanomachine’, a hypodermic needle-like structure (the injectisome) and the translocon, which forms a pore across the host cell membrane (Figure 1). Along with a combination of regulators and chaperones, the T3SS’s primary function is to inject multiple toxic Yersinia effector proteins (Yops) directly into the eukaryotic host cell cytosol. Once inside, they subvert host cell signalling pathways and trigger a pre-programmed metabolic chain reaction that results in apoptosis6,7. Yops also inhibit phagocytosis and block cytokine production.
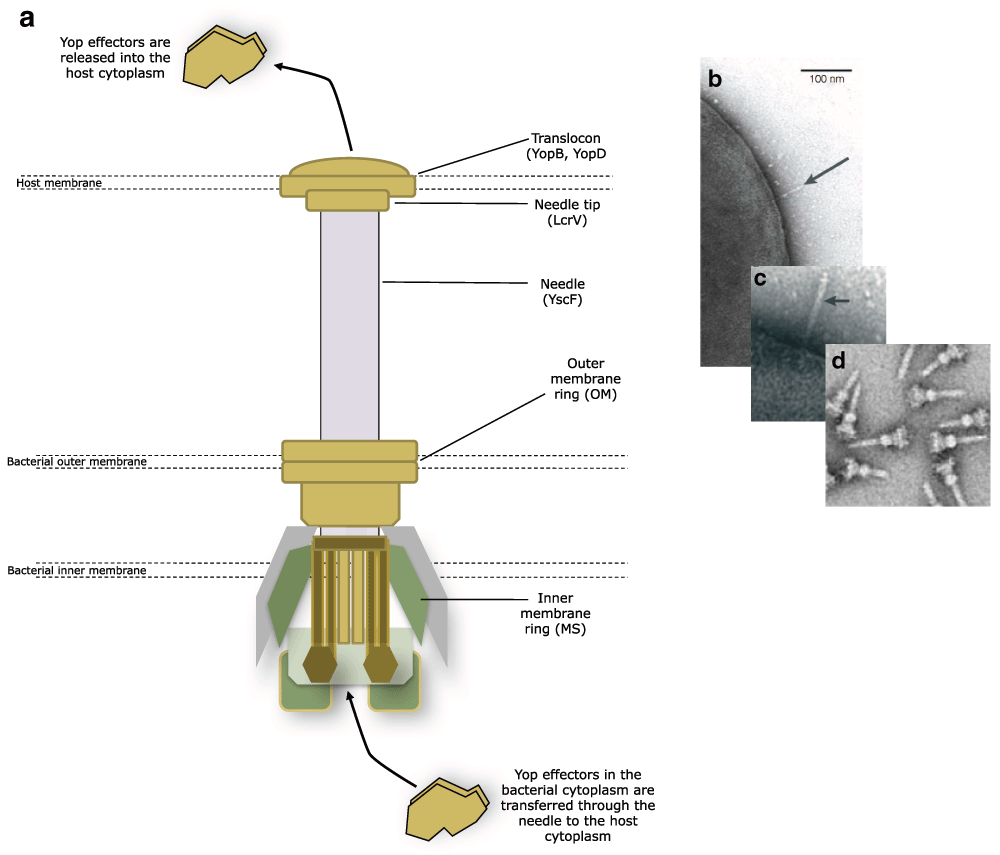
Figure 1. Assembly of the type three secretion system (T3SS) needle.
The needle is fixed into the bacterial inner and outer membrane and protrudes from the surface to penetrate the host membrane. The translocon forms a channel through the host membrane and the Yop effectors are transferred into the host from the bacterial cytoplasm via the needle and translocon (a). The needle protrudes from the bacterial surface prior to host cell penetration (b, c arrowed). Salmonella typhimurium T3SS needles isolated from the bacterial membrane (d). (a) adapted from 133, (b) reproduced with permission and taken from reference 56, (c) reproduced with permission and taken from reference 16, and (d) reproduced from reference 134.
The structure of the T3SS needle and translocon
Structurally, the base of the injectisome is composed of a number of proteins that adopt a cylindrical architecture similar to that of the flagellar basal body8 that are directed to the membrane by the secretion (Sec)-dependent pathway9. The injectisome incorporates two membrane rings termed the MS (membrane and supramembrane) and OM (outer membrane) rings. These are connected to five integral membrane proteins that play a role in exporting proteins10,11 (Figure 1). The export apparatus itself is flanked by YscQ, which facilitates the binding of the ATPase YscN and the secretion substrate-chaperone complexes12. YscN provides the proton motive force necessary for driving the secretion of the Yop effectors9,13,14.
Protruding into the extracellular space from the basal body is a hollow needle formed by the helical polymerisation of YscF protein subunits9,15,16. YscF is exported and polymerised in a T3SS-dependent manner along with YscP, a protein akin to a molecular ruler that determines the length of the needle and limits its size17–19. It has recently been shown that fully formed T3SS needles form clusters on the bacterial cell surface and new needles appear to localise to these clusters rather than being randomly distributed20 (Figure 2). The needle tip is capped with LcrV21,22, a protein that directs the formation of a pore or ‘translocon’23. The translocon consists of a tripartite protein pore, which is inserted into host cell membranes and drives the translocation of Yop effectors into the host target cell cytoplasm. The pore is composed of the transmembrane proteins YopB and YopD23 and the injectisome tip complex LcrV24–26. Bacteria lacking the tip and translocon proteins are able to secrete effectors into the extracellular environment but are defective in translocating Yops into host cells27–29.
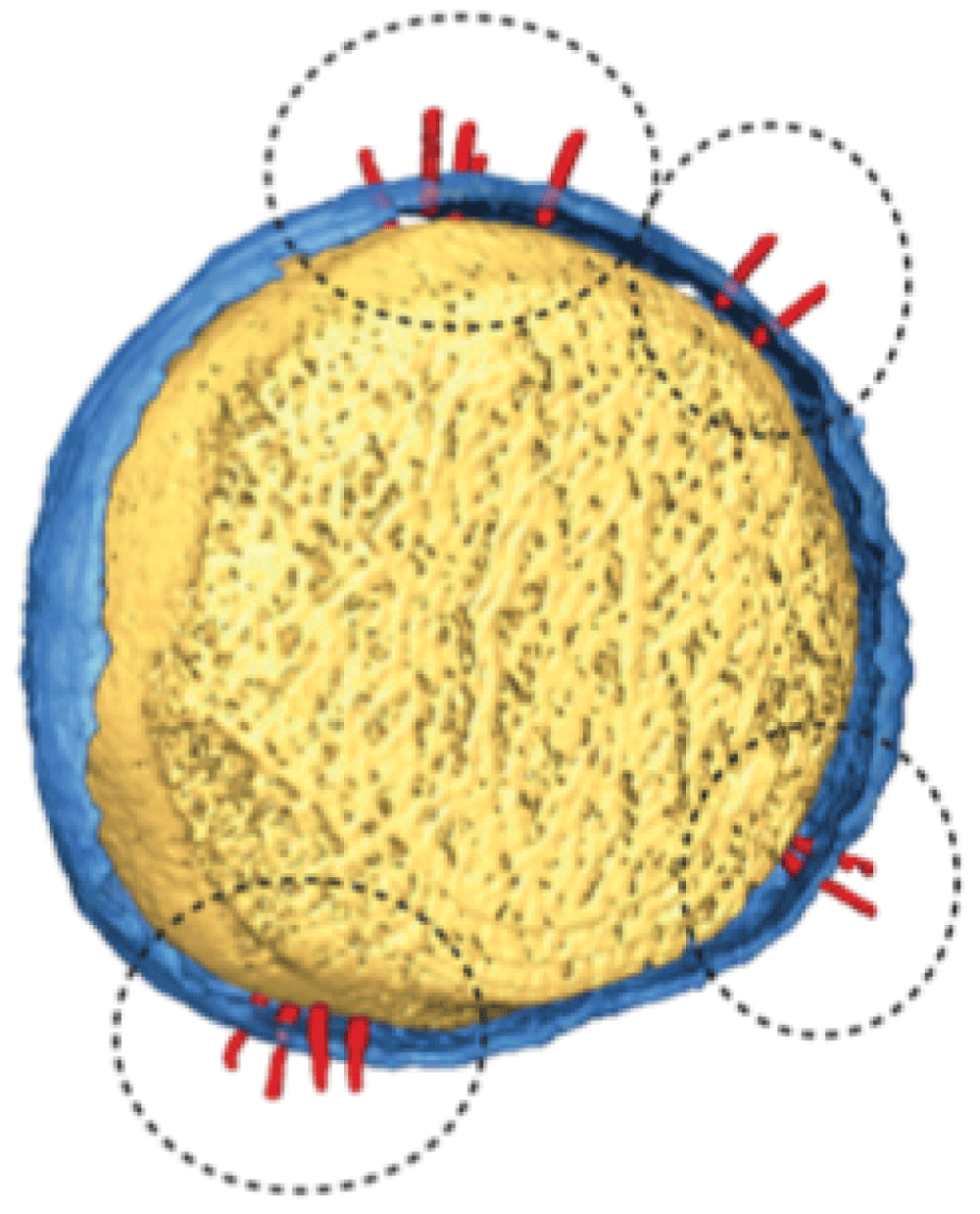
Figure 2. Type 3 secretion system (T3SS) needles (circled) appear to cluster together as they form at the cell surface.
Reproduced from reference 20.
Chaperones facilitate the formation and operation of the T3SS
Given the complexity of the T3SS, part of its sophistication relates to its in-built ability to discriminate between structural and secretion substrates, providing strict order to ensure the needle is assembled and polymerised before translocon and Yop effector secretion30. Such ordering requires specific chaperones, typically small protein dimers that protect the target T3SS protein from degradation31,32 and prevent premature oligomerisation24 and also ushering into the injectisome. These T3SS chaperones are usually subdivided into three classes: class I chaperones bind the Yop effector proteins and often share high structural conservation, class II chaperones associate with the translocon proteins YopB, YopD, and LcrV, and class III chaperones tend to form heterodimers and associate with structural components of the injectisome.
The Yop effectors
The Yop effector proteins are virulence factors synthesised in the bacterial cytoplasm and secreted through the T3SS needle and translocon into eukaryotic target cells (Figure 1). Four of these (YopE, YopT, YpkA, and YopH) are involved in disrupting the normal activities of the cytoskeleton and, apart from YopH, also target an important group of eukaryotic cell signalling components, the RhoA family of small GTPases that direct cytoskeletal rearrangements necessary for phagocytosis. YopE is a functional mimic of eukaryotic GTPase-activating proteins (GAPs)33 and disrupts the actin cytoskeleton34–36, resulting in the inhibition of phagocytosis by macrophages. YopT suppresses RhoA-mediated signalling by cleaving the post-translationally modified Rho GTPases37, which ultimately prevents the formation of the phagocytic cup for bacterial internalisation, and inhibits the assembly of focal adhesion complexes required for the development of pseudopodia and macrophage migration38,39. YpkA (YopO in Y. enterocolitica) associates with RhoA family proteins40,41 and inhibits phagocytosis42,43 by binding to and phosphorylating actin that is used as bait by Y. enterocolitica to titrate out host regulators responsible for actin polymerisation44. YopH is multi-functional and disrupts pathways involved in both innate and adaptive immunity and is essential for the virulence of Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica in mice45–47. YopH inhibits autophagy following binding of invasin or YadA (see next section) to β1-integrins48 and also blocks phagocytosis in macrophages49,50 by dephosphorylating focal adhesion complex proteins, which disrupts the link to the actin cytoskeleton51,52.
The remaining two effectors (YopJ and YopM) down-regulate elements of the immune system, such as inflammation and leukocyte recruitment53–57. YopJ (YopP in Y. enterocolitica) is a serine/threonine/lysine acetyltransferase that catalyses the acylation of kinases, inhibiting their ability to activate the release of NF-Κβ, which would otherwise induce pro-inflammatory cytokine production58–62. Recently, YopJ was also shown to play an important role in inhibiting caspase-1 in activated macrophages63.
YopM is translocated into macrophages64 and may also be able to self-deliver into some human cells65, yet it has no known enzymatic activity66 and its true function has yet to be elucidated. Inside eukaryotic cells, YopM may interact with and stimulate cellular kinases67 and is thought to localise to the nucleus68–70, where it may influence the expression of a range of genes, down-regulating many pro-inflammatory cytokines65,71, counteract the innate immune system by promoting depletion of natural killer cells in the liver, spleen, and blood72, and also prevent pyroptosis by binding to caspase-1, inhibiting its activity73,74.
Yersinia surface adhesins
For yersiniae to efficiently deliver Yops into the host, it is essential that they adhere to the host cell surface and remain in close association during the delivery process. To ensure that this is possible, the yersiniae produce virulence factors in addition to the T3SS. An active T3SS can deliver effector proteins into the host cell cytosol only if the bacterial cells make direct contact with, and bind tightly to, the host cell surface. Over the last 30 years, several chromosomally or plasmid-encoded protein virulence factors have been identified that play a variety of roles in host cell attachment prior to effector protein injection. In each case, attachment is not their exclusive function and not all are present or active in all three of the human pathogens. However, a combination of these proteins confers the ability to adhere to and invade host cells or bind sufficiently to ensure successful T3SS delivery of Yops.
Invasin
Invasin is a chromosomally encoded protein that mediates attachment to and entry into host cells by Y. pseudotuberculosis and Y. enterocolitica75, although in Y. pestis it is a pseudogene and therefore inactive76 (Figure 3). Invasin promotes small intestine epithelial cell internalisation by binding to host cell target receptors known as β1-integrins77 that present on the host cell surface. Integrins form clusters upon invasin binding, and the result is the rearrangement of the host cell cytoskeleton. This promotes phagocytosis and ultimately internalisation of the bacteria into the epithelial cells. In fact, invasin has a significantly greater (up to 100 times) affinity for some integrins than its natural ligand, fibronectin78, and such strong associations are believed to be major contributing factors to the efficiency of internalisation and Yop delivery into host cells.
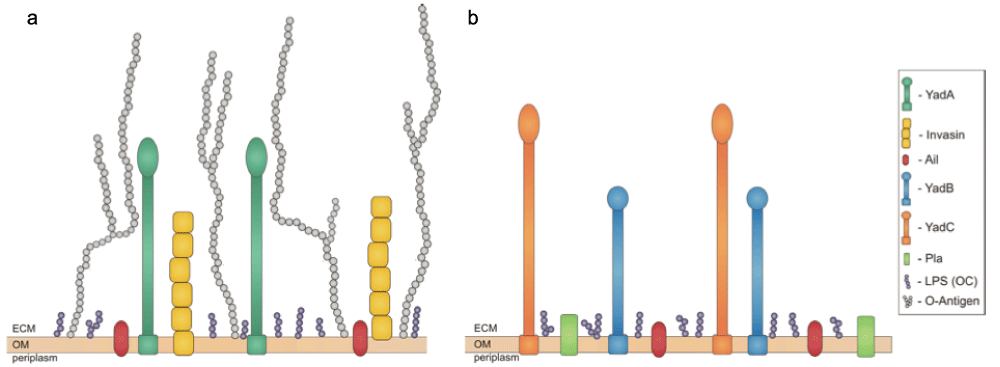
Figure 3.
Virulence factors found on the surface of Yersinia pseudotuberculosis, Yersinia enterocolitica (a), and Yersinia pestis (b) Ail, YadB, and YadC are shared by all three pathogens – YadB and YadC are absent from panel (a) for clarity – while Pla is unique to Y. pestis. YadA and invasin are important adhesins in Y. pseudotuberculosis and Y. enterocolitica but are not expressed by Y. pestis. Reproduced from reference 95.
Invasin expression is regulated by both temperature and pH in Y. enterocolitica79,80. The invasin gene is maximally expressed at 26°C, peaking during late exponential/early stationary phase with lower expression levels observed at 37°C. This apparent contradiction, since invasin is required for infection at 37°C, was resolved when Pepe et al. revealed that the expression of invasin at 37°C was restored to levels seen at 26°C when the pH was reduced to 5.5. It has been suggested that rather than an experimental artefact, the expression of invasin at ambient temperatures could prepare the bacteria for infection following ingestion and promote rapid transcytosis through the epithelia81,82. The pH effect is not evident in Y. pseudotuberculosis, suggesting that the mechanisms of regulation of invasin expression may differ between the two species83. Two regulators have been found to be important for invasin expression: RovA, required for the positive regulation of invasin, and YmoA, required for negative regulation83–85. Both RovA and YmoA recognise the promoter region of invasin and compete for binding. Once RovA is bound, it appears to prevent YmoA from binding, thus inhibiting negative regulation of invasin86,87. The expression of rovA is itself regulated by temperature via RovM, which acts as a repressor of rovA expression under inducing growth conditions88.
YadA
After crossing the intestinal epithelium, the major adhesin responsible for Yersinia contact with cells of the submucosa is the virulence plasmid-encoded protein YadA (recently reviewed by Mühlenkamp et al.89) (Figure 3). YadA expression is induced at or above 37°C90,91, and under these conditions it is so abundant that it can virtually coat the entire outer surface of the bacterial cell92. Interestingly, despite YadA’s utility and abundance, Y. pestis possesses an inactive yadA pseudogene due to a single nucleotide deletion that results in a frame-shift mutation93 (Figure 3). Although Y. pestis does not produce a functional YadA protein, the chromosome carries two orthologues, YadB and YadC. Also found in Y. pseudotuberculosis, these two proteins are not thought to play a role in adherence but may contribute to host cell invasion. They may also be required for full virulence and lethality in bubonic but not pneumonic plague in mouse infection models94.
YadA is a non-fimbrial adhesin95 belonging to the trimeric autotransporter adhesin family members, which are usually referred to as obligate homotrimeric proteins. The protein is shaped like a lollipop, with an N-terminal globular head domain connected by a coiled-coil stalk to a C-terminal anchor domain embedded in the outer membrane92. YadA has multiple functions but as an adhesin may act as a docking system, allowing the injectisome of the T3SS to come into contact with the target cell membrane to deliver the Yop effector proteins96.
Until recently, it was thought that YadA bound only to the large proteins of the extracellular matrix – collagen, fibronectin, and laminin – which in turn bind β1-integrins97–99. However, Keller et al.100 recently discovered that YadA-mediated adhesion may be facilitated by a broad range of host cell receptors and in the absence of β1-integrins may facilitate Yop injection via αV integrins as well as other unidentified cofactors. Y. enterocolitica YadA also binds leukocytes in a β1-integrin-independent manner during systemic infection101, all of which suggests that YadA has the potential to target a broad range of cell types to ensure efficient Yop delivery.
The collagen-binding activity of YadA in Y. enterocolitica is an absolute requirement for pathogenicity; however, YadA is not essential for virulence in Y. pseudotuberculosis97. YadA mediates adhesion to a number of cell types, including epithelial cells and macrophages, and can act as a haemagglutinin97. In Y. pseudotuberculosis, YadA promotes the invasion of epithelial cells and is interchangeable with the activity of invasin102, although Y. enterocolitica YadA is not as efficient an invasin as that of Y. pseudotuberculosis103. YadA also mediates bacteria-bacteria autoagglutination, since the head domain has an affinity for itself92. This self-affinity also promotes the formation of densely packed microcolonies that may promote antiphagocytic activity in Y. enterocolitica. YadA also binds to intestinal mucus104 and plays a major role in conferring serum resistance105–107.
Ail
The ail locus is chromosomally located and encodes a 17 kDa surface-associated protein (Figure 3) that is thermally regulated, being maximally expressed at 37°C108,109. In Y. enterocolitica, Ail-directed adhesion to host cells shows more specificity than invasin, as it allows invasion of some cell lines, such as human laryngeal epithelial type 2 (HEp-2), human endometrial (HEC-1B), and Chinese hamster ovary (CHO) cells, but no invasion of Madin-Darby canine kidney (MDCK) cells110. Both laminin and fibronectin are known targets for Y. pestis Ail binding111 and vitronectin is actively recruited to the Y. pestis surface through the activities of Ail112. Interestingly, Y. pseudotuberculosis Ail is unable to promote the attachment and invasion phenotypes when expressed in Escherichia coli113. As with invasin, Ail-mediated tight attachment to host cells presumably ensures that Yop delivery is rapid and efficient114. Aside from its adhesive properties, Ail also confers resistance to serum killing115 in all three human pathogenic yersiniae. It is apparent that Ail plays a more prominent role in the virulence of Y. pestis, which is presumably owing to the fact that the other prominent virulence factors contributing many of the Ail functions in Y. pseudotuberculosis and Y. enterocolitica are dysfunctional.
Psa – the pH6 antigen
The chromosomally encoded pH6 antigen (Psa) was originally identified in Y. pestis as a surface antigen expressed at mammalian body temperatures at pH values similar to those found in phagolysosomes116. It was subsequently found to cause the agglutination of erythrocytes113,117. Further investigation revealed a cell surface complex composed of aggregates of a 15 kDa protein (PsaA) that requires two regulators, PsaE and PsaF, for maximal induction118,119. PsaA possesses a flexible fimbrial structure that is highly expressed during macrophage infection120. Biochemical examination of Psa reveals that it binds to β1-linked galactosyl residues in glycosphingolipids121, mainly of the type found in apolipoprotein-B-containing lipoproteins in human plasma, such as low-density lipoprotein (LDL) and in lipid rafts in macrophage membranes122. Furthermore, Psa acts as a bacterial Fc receptor, binding human immunoglobulin (IgG) but not reacting with rabbit, mouse, or sheep IgG123. As with the other adhesins, the activities of Psa appear to mediate Yop secretion. Y. pseudotuberculosis and Y. enterocolitica both produce a surface protein analogous to Psa but it is referred to as MyfA. Both Psa and MyfA coat the bacterial surface with a fibrillar matrix120,124 and in Y. pseudotuberculosis MyfA promotes attachment to tissue culture cells and haemagglutination113.
Y. pestis plasmid-specific virulence factors
Apart from the T3SS virulence plasmid, two other plasmids, pPCP and pMT (sometimes referred to as pFra) that are unique to Y. pestis, possess additional virulence factors. pPCP encodes the plasminogen activator Pla protease/adhesin (Figure 3). Pla converts plasminogen to plasmin125,126, which then degrades extracellular matrices and confers on Y. pestis the ability to rapidly invade the host and migrate to lymphatic tissues127,128. The over-activation of plasmin results in laminin and fibrin clot degradation, exacerbating migration across host barriers129, which is further compounded by the activities of Pla as an adhesin and an invasin130,131. pMT is responsible for the production of a murine toxin that is required during the colonisation of fleas132.
Concluding remarks
Over the last three decades, a considerable amount of detailed knowledge has accumulated that has enabled us to understand how the yersiniae colonise tissues and combat host defences during infection. While the Yersinia T3SS is perhaps the best understood system of its kind, many questions remain unanswered. For example, fully elucidating the function of YopM will offer an important step change, as will understanding more clearly the global molecular mechanisms that underpin the regulatory relationships that must exist between the T3SS system and the adhesins. It is also important to try to understand the relationships that exist between the different adhesins, how they compensate for each other, and which environmental signals dictate their site-specific expression. Finally, although the structures of many of the adhesins have been elucidated, there is certainly a need to better understand how they interact with different host ligands. While significant progress has been made in defining this sophisticated and finely tuned arsenal of virulence determinants, much more work is required to fully appreciate the success of the yersiniae as pathogens.
Competing interests
The authors declare that they have no competing interests.
Grant information
The author(s) declared that no grants were involved in supporting this work.
Faculty Opinions recommendedReferences
- 1.
Achtman M, Zurth K, Morelli G, et al.:
Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis.
Proc Natl Acad Sci U S A.
1999; 96(24): 14043–8. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 2.
Achtman M, Morelli G, Zhu P, et al.:
Microevolution and history of the plague bacillus, Yersinia pestis.
Proc Natl Acad Sci U S A.
2004; 101(51): 17837–42. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 3.
Prentice MB, Rahalison L:
Plague.
Lancet.
2007; 369(9568): 1196–207. PubMed Abstract
| Publisher Full Text
- 4.
Stenseth NC, Atshabar BB, Begon M, et al.:
Plague: past, present, and future.
PLoS Med.
2008; 5(1): e3. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 5.
Guiyoule A, Gerbaud G, Buchrieser C, et al.:
Transferable plasmid-mediated resistance to streptomycin in a clinical isolate of Yersinia pestis.
Emerg Infect Dis.
2001; 7(1): 43–8. PubMed Abstract
| Free Full Text
- 6.
Wren BW:
The yersiniae--a model genus to study the rapid evolution of bacterial pathogens.
Nat Rev Microbiol.
2003; 1(1): 55–64. PubMed Abstract
| Publisher Full Text
- 7.
Trosky JE, Liverman AD, Orth K:
Yersinia outer proteins: Yops.
Cell Microbiol.
2008; 10(3): 557–65. PubMed Abstract
| Publisher Full Text
- 8.
Diepold A, Armitage JP:
Type III secretion systems: the bacterial flagellum and the injectisome.
Philos Trans R Soc Lond, B Biol Sci.
2015; 370(1679): pii: 20150020. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 9.
Diepold A, Amstutz M, Abel S, et al.:
Deciphering the assembly of the Yersinia type III secretion injectisome.
EMBO J.
2010; 29(11): 1928–40. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 10.
Edqvist PJ, Olsson J, Lavander M, et al.:
YscP and YscU regulate substrate specificity of the Yersinia type III secretion system.
J Bacteriol.
2003; 185(7): 2259–66. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 11.
Sorg I, Wagner S, Amstutz M, et al.:
YscU recognizes translocators as export substrates of the Yersinia injectisome.
EMBO J.
2007; 26(12): 3015–24. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 12.
Jackson MW, Plano GV:
Interactions between type III secretion apparatus components from Yersinia pestis detected using the yeast two-hybrid system.
FEMS Microbiol Lett.
2000; 186(1): 85–90. PubMed Abstract
| Publisher Full Text
- 13.
Zarivach R, Vuckovic M, Deng W, et al.:
Structural analysis of a prototypical ATPase from the type III secretion system.
Nat Struct Mol Biol.
2007; 14(2): 131–7. PubMed Abstract
| Publisher Full Text
- 14.
Woestyn S, Allaoui A, Wattiau P, et al.:
YscN, the putative energizer of the Yersinia Yop secretion machinery.
J Bacteriol.
1994; 176(6): 1561–9. PubMed Abstract
| Free Full Text
- 15.
Hoiczyk E, Blobel G:
Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells.
Proc Natl Acad Sci U S A.
2001; 98(8): 4669–74. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 16.
Cornelis GR:
The type III secretion injectisome.
Nat Rev Microbiol.
2006; 4(11): 811–25. PubMed Abstract
| Publisher Full Text
- 17.
Stainier I, Bleves S, Josenhans C, et al.:
YscP, a Yersinia protein required for Yop secretion that is surface exposed, and released in low Ca2+.
Mol Microbiol.
2000; 37(5): 1005–18. PubMed Abstract
| Publisher Full Text
- 18.
Wagner S, Sorg I, Degiacomi M, et al.:
The helical content of the YscP molecular ruler determines the length of the Yersinia injectisome.
Mol Microbiol.
2009; 71(3): 692–701. PubMed Abstract
| Publisher Full Text
- 19.
Payne PL, Straley SC:
YscP of Yersinia pestis is a secreted component of the Yop secretion system.
J Bacteriol.
1999; 181(9): 2852–62. PubMed Abstract
| Free Full Text
- 20.
Kudryashev M, Diepold A, Amstutz M, et al.:
Yersinia enterocolitica type III secretion injectisomes form regularly spaced clusters, which incorporate new machines upon activation.
Mol Microbiol.
2015; 95(5): 875–84. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 21.
Broz P, Mueller CA, Müller SA, et al.:
Function and molecular architecture of the Yersinia injectisome tip complex.
Mol Microbiol.
2007; 65(5): 1311–20. PubMed Abstract
| Publisher Full Text
- 22.
Mueller CA, Broz P, Müller SA, et al.:
The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles.
Science.
2005; 310(5748): 674–6. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 23.
Håkansson S, Bergman T, Vanooteghem JC, et al.:
YopB and YopD constitute a novel class of Yersinia Yop proteins.
Infect Immun.
1993; 61(1): 71–80. PubMed Abstract
| Free Full Text
- 24.
Neyt C, Cornelis GR:
Role of SycD, the chaperone of the Yersinia Yop translocators YopB and YopD.
Mol Microbiol.
1999; 31(1): 143–56. PubMed Abstract
| Publisher Full Text
- 25.
Bröms JE, Forslund AL, Forsberg A, et al.:
Dissection of homologous translocon operons reveals a distinct role for YopD in type III secretion by Yersinia pseudotuberculosis.
Microbiology.
2003; 149(Pt 9): 2615–26. PubMed Abstract
| Publisher Full Text
- 26.
Holmström A, Olsson J, Cherepanov P, et al.:
LcrV is a channel size-determining component of the Yop effector translocon of Yersinia.
Mol Microbiol.
2001; 39(3): 620–32. PubMed Abstract
| Publisher Full Text
- 27.
DeBord KL, Lee VT, Schneewind O:
Roles of LcrG and LcrV during type III targeting of effector Yops by Yersinia enterocolitica.
J Bacteriol.
2001; 183(15): 4588–98. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 28.
Lee VT, Schneewind O:
Type III machines of pathogenic yersiniae secrete virulence factors into the extracellular milieu.
Mol Microbiol.
1999; 31(6): 1619–29. PubMed Abstract
| Publisher Full Text
- 29.
Cheng LW, Schneewind O:
Yersinia enterocolitica TyeA, an intracellular regulator of the type III machinery, is required for specific targeting of YopE, YopH, YopM, and YopN into the cytosol of eukaryotic cells.
J Bacteriol.
2000; 182(11): 3183–90. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 30.
Cornelis GR, Wolf-Watz H:
The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells.
Mol Microbiol.
1997; 23(5): 861–7. PubMed Abstract
| Publisher Full Text
- 31.
Woestyn S, Sory MP, Boland A, et al.:
The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes.
Mol Microbiol.
1996; 20(6): 1261–71. PubMed Abstract
| Publisher Full Text
- 32.
Cheng LW, Schneewind O:
Yersinia enterocolitica type III secretion. On the role of SycE in targeting YopE into HeLa cells.
J Biol Chem.
1999; 274(31): 22102–8. PubMed Abstract
| Publisher Full Text
- 33.
Evdokimov AG, Tropea JE, Routzahn KM, et al.:
Crystal structure of the Yersinia pestis GTPase activator YopE.
Protein Sci.
2002; 11(2): 401–8. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 34.
Black DS, Bliska JB:
The RhoGAP activity of the Yersinia pseudotuberculosis cytotoxin YopE is required for antiphagocytic function and virulence.
Mol Microbiol.
2000; 37(3): 515–27. PubMed Abstract
| Publisher Full Text
- 35.
Aili M, Isaksson EL, Hallberg B, et al.:
Functional analysis of the YopE GTPase-activating protein (GAP) activity of Yersinia pseudotuberculosis.
Cell Microbiol.
2006; 8(6): 1020–33. PubMed Abstract
| Publisher Full Text
- 36.
Andor A, Trülzsch K, Essler M, et al.:
YopE of Yersinia, a GAP for Rho GTPases, selectively modulates Rac-dependent actin structures in endothelial cells.
Cell Microbiol.
2001; 3(5): 301–10. PubMed Abstract
| Publisher Full Text
- 37.
Shao F, Merritt PM, Bao Z, et al.:
A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis.
Cell.
2002; 109(5): 575–88. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 38.
Aepfelbacher M, Trasak C, Wiedemann A, et al.:
Rho-GTP binding proteins in Yersinia target cell interaction.
Adv Exp Med Biol.
2003; 529: 65–72. PubMed Abstract
| Publisher Full Text
- 39.
Schmidt G:
Yersinia enterocolitica outer protein T (YopT).
Eur J Cell Biol.
2011; 90(11): 955–8. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 40.
Barz C, Abahji TN, Trülzsch K, et al.:
The Yersinia Ser/Thr protein kinase YpkA/YopO directly interacts with the small GTPases RhoA and Rac-1.
FEBS Lett.
2000; 482(1–2): 139–43. PubMed Abstract
| Publisher Full Text
- 41.
Groves E, Rittinger K, Amstutz M, et al.:
Sequestering of Rac by the Yersinia effector YopO blocks Fcgamma receptor-mediated phagocytosis.
J Biol Chem.
2010; 285(6): 4087–98. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 42.
Navarro L, Koller A, Nordfelth R, et al.:
Identification of a molecular target for the Yersinia protein kinase A.
Mol Cell.
2007; 26(4): 465–77. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 43.
Juris SJ, Rudolph AE, Huddler D, et al.:
A distinctive role for the Yersinia protein kinase: actin binding, kinase activation, and cytoskeleton disruption.
Proc Natl Acad Sci U S A.
2000; 97(17): 9431–6. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 44.
Lee WL, Grimes JM, Robinson RC:
Yersinia effector YopO uses actin as bait to phosphorylate proteins that regulate actin polymerization.
Nat Struct Mol Biol.
2015; 22(3): 248–55. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 45.
Cantwell AM, Bubeck SS, Dube PH:
YopH inhibits early pro-inflammatory cytokine responses during plague pneumonia.
BMC Immunol.
2010; 11: 29. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 46.
Logsdon LK, Mecsas J:
Requirement of the Yersinia pseudotuberculosis effectors YopH and YopE in colonization and persistence in intestinal and lymph tissues.
Infect Immun.
2003; 71(8): 4595–607. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 47.
Trülzsch K, Sporleder T, Igwe EI, et al.:
Contribution of the major secreted yops of Yersinia enterocolitica O:8 to pathogenicity in the mouse infection model.
Infect Immun.
2004; 72(9): 5227–34. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 48.
Deuretzbacher A, Czymmeck N, Reimer R, et al.:
Beta1 integrin-dependent engulfment of Yersinia enterocolitica by macrophages is coupled to the activation of autophagy and suppressed by type III protein secretion.
J Immunol.
2009; 183(9): 5847–60. PubMed Abstract
| Publisher Full Text
- 49.
Rosqvist R, Skurnik M, Wolf-Watz H:
Increased virulence of Yersinia pseudotuberculosis by two independent mutations.
Nature.
1988; 334(6182): 522–4. PubMed Abstract
| Publisher Full Text
- 50.
Bölin I, Wolf-Watz H:
The plasmid-encoded Yop2b protein of Yersinia pseudotuberculosis is a virulence determinant regulated by calcium and temperature at the level of transcription.
Mol Microbiol.
1988; 2(2): 237–45. PubMed Abstract
| Publisher Full Text
- 51.
Bliska JB, Guan KL, Dixon JE, et al.:
Tyrosine phosphate hydrolysis of host proteins by an essential Yersinia virulence determinant.
Proc Natl Acad Sci U S A.
1991; 88(4): 1187–91. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 52.
Navarro L, Alto NM, Dixon JE:
Functions of the Yersinia effector proteins in inhibiting host immune responses.
Curr Opin Microbiol.
2005; 8(1): 21–7. PubMed Abstract
| Publisher Full Text
- 53.
Grosdent N, Maridonneau-Parini I, Sory MP, et al.:
Role of Yops and adhesins in resistance of Yersinia enterocolitica to phagocytosis.
Infect Immun.
2002; 70(8): 4165–76. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 54.
Shao F:
Biochemical functions of Yersinia type III effectors.
Curr Opin Microbiol.
2008; 11(1): 21–9. PubMed Abstract
| Publisher Full Text
- 55.
Matsumoto H, Young GM:
Translocated effectors of Yersinia.
Curr Opin Microbiol.
2009; 12(1): 94–100. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 56.
Cornelis GR:
The Yersinia Ysc-Yop 'type III' weaponry.
Nat Rev Mol Cell Biol.
2002; 3(10): 742–52. PubMed Abstract
| Publisher Full Text
- 57.
Cornelis GR:
Yersinia type III secretion: send in the effectors.
J Cell Biol.
2002; 158(3): 401–8. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 58.
Mukherjee S, Keitany G, Li Y, et al.:
Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation.
Science.
2006; 312(5777): 1211–4. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 59.
Orth K, Xu Z, Mudgett MB, et al.:
Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease.
Science.
2000; 290(5496): 1594–7. PubMed Abstract
| Publisher Full Text
- 60.
Mittal R, Peak-Chew SY, McMahon HT:
Acetylation of MEK2 and I kappa B kinase (IKK) activation loop residues by YopJ inhibits signaling.
Proc Natl Acad Sci U S A.
2006; 103(49): 18574–9. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 61.
Ruckdeschel K, Harb S, Roggenkamp A, et al.:
Yersinia enterocolitica impairs activation of transcription factor NF-kappaB: involvement in the induction of programmed cell death and in the suppression of the macrophage tumor necrosis factor alpha production.
J Exp Med.
1998; 187(7): 1069–79. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 62.
Boland A, Cornelis GR:
Role of YopP in suppression of tumor necrosis factor alpha release by macrophages during Yersinia infection.
Infect Immun.
1998; 66(5): 1878–84. PubMed Abstract
| Free Full Text
- 63.
Schoberle TJ, Chung LK, McPhee JB, et al.:
Uncovering an Important Role for YopJ in the Inhibition of Caspase-1 in Activated Macrophages and Promoting Yersinia pseudotuberculosis Virulence.
Infect Immun.
2016; 84(4): 1062–72. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 64.
Boland A, Sory MP, Iriarte M, et al.:
Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y.enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus.
EMBO J.
1996; 15(19): 5191–201. PubMed Abstract
| Free Full Text
- 65.
Bertrand J, Ruther C, Scharnert J, et al.:
The bacterial effector protein YopM is a selfdelivering immune therapeutic agent that reduces inflammation in rheumatoid arthritis.
Ann Rheum Dis.
2010; 69: A47. Publisher Full Text
| Faculty Opinions Recommendation
- 66.
Höfling S, Grabowski B, Norkowski S, et al.:
Current activities of the Yersinia effector protein YopM.
Int J Med Microbiol.
2015; 305(3): 424–32. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 67.
Hentschke M, Berneking L, Belmar Campos C, et al.:
Yersinia virulence factor YopM induces sustained RSK activation by interfering with dephosphorylation.
PLoS One.
2010; 5(10): pii: e13165. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 68.
Benabdillah R, Mota LJ, Lützelschwab S, et al.:
Identification of a nuclear targeting signal in YopM from Yersinia spp.
Microb Pathog.
2004; 36(5): 247–61. PubMed Abstract
| Publisher Full Text
- 69.
Skrzypek E, Myers-Morales T, Whiteheart SW, et al.:
Application of a Saccharomyces cerevisiae model to study requirements for trafficking of Yersinia pestis YopM in eucaryotic cells.
Infect Immun.
2003; 71(2): 937–47. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 70.
Skrzypek E, Cowan C, Straley SC:
Targeting of the Yersinia pestis YopM protein into HeLa cells and intracellular trafficking to the nucleus.
Mol Microbiol.
1998; 30(5): 1051–65. PubMed Abstract
| Publisher Full Text
- 71.
Ye Z, Kerschen EJ, Cohen DA, et al.:
Gr1+ cells control growth of YopM-negative yersinia pestis during systemic plague.
Infect Immun.
2009; 77(9): 3791–806. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 72.
Kerschen EJ, Cohen DA, Kaplan AM, et al.:
The plague virulence protein YopM targets the innate immune response by causing a global depletion of NK cells.
Infect Immun.
2004; 72(8): 4589–602. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 73.
LaRock CN, Cookson BT:
The Yersinia virulence effector YopM binds caspase-1 to arrest inflammasome assembly and processing.
Cell Host Microbe.
2012; 12(6): 799–805. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 74.
Chung LK, Philip NH, Schmidt VA, et al.:
IQGAP1 is important for activation of caspase-1 in macrophages and is targeted by Yersinia pestis type III effector YopM.
MBio.
2014; 5(4): e01402–14. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 75.
Isberg RR, Voorhis DL, Falkow S:
Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells.
Cell.
1987; 50(5): 769–78. PubMed Abstract
| Publisher Full Text
- 76.
Parkhill J, Wren BW, Thomson NR, et al.:
Genome sequence of Yersinia pestis, the causative agent of plague.
Nature.
2001; 413(6855): 523–7. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 77.
Clark MA, Hirst BH, Jepson MA:
M-cell surface beta1 integrin expression and invasin-mediated targeting of Yersinia pseudotuberculosis to mouse Peyer's patch M cells.
Infect Immun.
1998; 66(3): 1237–43. PubMed Abstract
| Free Full Text
- 78.
Hamburger ZA, Brown MS, Isberg RR, et al.:
Crystal structure of invasin: a bacterial integrin-binding protein.
Science.
1999; 286(5438): 291–5. PubMed Abstract
| Publisher Full Text
- 79.
Isberg RR, Swain A, Falkow S:
Analysis of expression and thermoregulation of the Yersinia pseudotuberculosis inv gene with hybrid proteins.
Infect Immun.
1988; 56(8): 2133–8. PubMed Abstract
| Free Full Text
- 80.
Pepe JC, Badger JL, Miller VL:
Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene.
Mol Microbiol.
1994; 11(1): 123–35. PubMed Abstract
| Publisher Full Text
- 81.
Grassl GA, Bohn E, Müller Y, et al.:
Interaction of Yersinia enterocolitica with epithelial cells: invasin beyond invasion.
Int J Med Microbiol.
2003; 293(1): 41–54. PubMed Abstract
| Publisher Full Text
- 82.
Uliczka F, Pisano F, Kochut A, et al.:
Monitoring of gene expression in bacteria during infections using an adaptable set of bioluminescent, fluorescent and colorigenic fusion vectors.
PLoS One.
2011; 6(6): e20425. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 83.
Nagel G, Lahrz A, Dersch P:
Environmental control of invasin expression in Yersinia pseudotuberculosis is mediated by regulation of RovA, a transcriptional activator of the SlyA/Hor family.
Mol Microbiol.
2001; 41(6): 1249–6. PubMed Abstract
| Publisher Full Text
- 84.
Revell PA, Miller VL:
A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence.
Mol Microbiol.
2000; 35(3): 677–85. PubMed Abstract
| Publisher Full Text
- 85.
Ellison DW, Young B, Nelson K, et al.:
YmoA negatively regulates expression of invasin from Yersinia enterocolitica.
J Bacteriol.
2003; 185(24): 7153–9. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 86.
Heroven AK, Nagel G, Tran HJ, et al.:
RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis.
Mol Microbiol.
2004; 53(3): 871–88. PubMed Abstract
| Publisher Full Text
- 87.
Ellison DW, Miller VL:
H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA.
J Bacteriol.
2006; 188(14): 5101–12. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 88.
Heroven AK, Dersch P:
RovM, a novel LysR-type regulator of the virulence activator gene rovA, controls cell invasion, virulence and motility of Yersinia pseudotuberculosis.
Mol Microbiol.
2006; 62(5): 1469–83. PubMed Abstract
| Publisher Full Text
- 89.
Mühlenkamp M, Oberhettinger P, Leo JC, et al.:
Yersinia adhesin A (YadA)--beauty & beast.
Int J Med Microbiol.
2015; 305(2): 252–8. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 90.
Skurnik M, Toivanen P:
LcrF is the temperature-regulated activator of the yadA gene of Yersinia enterocolitica and Yersinia pseudotuberculosis.
J Bacteriol.
1992; 174(6): 2047–51. PubMed Abstract
| Free Full Text
- 91.
Zaleska M, Lounatmaa K, Nurminen M, et al.:
A novel virulence-associated cell surface structure composed of 47-kd protein subunits in Yersinia enterocolitica.
EMBO J.
1985; 4(4): 1013–8. PubMed Abstract
| Free Full Text
- 92.
Hoiczyk E, Roggenkamp A, Reichenbecher M, et al.:
Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins.
EMBO J.
2000; 19(22): 5989–99. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 93.
Skurnik M, Wolf-Watz H:
Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp.
Mol Microbiol.
1989; 3(4): 517–29. PubMed Abstract
| Publisher Full Text
- 94.
Forman S, Wulff CR, Myers-Morales T, et al.:
yadBC of Yersinia pestis, a new virulence determinant for bubonic plague.
Infect Immun.
2008; 76(2): 578–87. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 95.
Mikula KM, Kolodziejczyk R, Goldman A:
Yersinia infection tools-characterization of structure and function of adhesins.
Front Cell Infect Microbiol.
2013; 2: 169. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 96.
Visser LG, Annema A, van Furth R:
Role of Yops in inhibition of phagocytosis and killing of opsonized Yersinia enterocolitica by human granulocytes.
Infect Immun.
1995; 63(7): 2570–5. PubMed Abstract
| Free Full Text
- 97.
El Tahir Y, Skurnik M:
YadA, the multifaceted Yersinia adhesin.
Int J Med Microbiol.
2001; 291(3): 209–18. PubMed Abstract
| Publisher Full Text
- 98.
Tertti R, Skurnik M, Vartio T, et al.:
Adhesion protein YadA of Yersinia species mediates binding of bacteria to fibronectin.
Infect Immun.
1992; 60(7): 3021–4. PubMed Abstract
| Free Full Text
- 99.
Flügel A, Schulze-Koops H, Heesemann J, et al.:
Interaction of enteropathogenic Yersinia enterocolitica with complex basement membranes and the extracellular matrix proteins collagen type IV, laminin-1 and -2, and nidogen/entactin.
J Biol Chem.
1994; 269(47): 29732–8. PubMed Abstract
- 100.
Keller B, Mühlenkamp M, Deuschle E, et al.:
Yersinia enterocolitica exploits different pathways to accomplish adhesion and toxin injection into host cells.
Cell Microbiol.
2015; 17(8): 1179–204. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 101.
Deuschle E, Keller B, Siegfried A, et al.:
Role of β1 integrins and bacterial adhesins for Yop injection into leukocytes in Yersinia enterocolitica systemic mouse infection.
Int J Med Microbiol.
2016; 306(2): 77–88. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 102.
Eitel J, Dersch P:
The YadA protein of Yersinia pseudotuberculosis mediates high-efficiency uptake into human cells under environmental conditions in which invasin is repressed.
Infect Immun.
2002; 70(9): 4880–91. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 103.
Heise T, Dersch P:
Identification of a domain in Yersinia virulence factor YadA that is crucial for extracellular matrix-specific cell adhesion and uptake.
Proc Natl Acad Sci U S A.
2006; 103(9): 3375–80. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 104.
Mantle M, Husar SD:
Binding of Yersinia enterocolitica to purified, native small intestinal mucins from rabbits and humans involves interactions with the mucin carbohydrate moiety.
Infect Immun.
1994; 62(4): 1219–27. PubMed Abstract
| Free Full Text
- 105.
Biedzka-Sarek M, Jarva H, Hyytiäinen H, et al.:
Characterization of complement factor H binding to Yersinia enterocolitica serotype O:3.
Infect Immun.
2008; 76(9): 4100–9. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 106.
Biedzka-Sarek M, Salmenlinna S, Gruber M, et al.:
Functional mapping of YadA- and Ail-mediated binding of human factor H to Yersinia enterocolitica serotype O:3.
Infect Immun.
2008; 76(11): 5016–27. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 107.
Kirjavainen V, Jarva H, Biedzka-Sarek M, et al.:
Yersinia enterocolitica serum resistance proteins YadA and ail bind the complement regulator C4b-binding protein.
PLoS Pathog.
2008; 4(8): e1000140. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 108.
Miller VL, Falkow S:
Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells.
Infect Immun.
1988; 56(5): 1242–8. PubMed Abstract
| Free Full Text
- 109.
Miller VL, Bliska JB, Falkow S:
Nucleotide sequence of the Yersinia enterocolitica ail gene and characterization of the Ail protein product.
J Bacteriol.
1990; 172(2): 1062–9. PubMed Abstract
| Free Full Text
- 110.
Miller VL, Mekalanos JJ:
A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR.
J Bacteriol.
1988; 170(6): 2575–83. PubMed Abstract
| Free Full Text
- 111.
Yamashita S, Lukacik P, Barnard TJ, et al.:
Structural insights into Ail-mediated adhesion in Yersinia pestis.
Structure.
2011; 19(11): 1672–82. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 112.
Bartra SS, Ding Y, Fujimoto LM, et al.:
Yersinia pestis uses the Ail outer membrane protein to recruit vitronectin.
Microbiology.
2015; 161(11): 2174–83. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 113.
Yang Y, Merriam JJ, Mueller JP, et al.:
The psa locus is responsible for thermoinducible binding of Yersinia pseudotuberculosis to cultured cells.
Infect Immun.
1996; 64(7): 2483–9. PubMed Abstract
| Free Full Text
- 114.
Felek S, Muszyński A, Carlson RW, et al.:
Phosphoglucomutase of Yersinia pestis is required for autoaggregation and polymyxin B resistance.
Infect Immun.
2010; 78(3): 1163–75. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 115.
Pierson DE, Falkow S:
The ail gene of Yersinia enterocolitica has a role in the ability of the organism to survive serum killing.
Infect Immun.
1993; 61(5): 1846–52. PubMed Abstract
| Free Full Text
- 116.
Ben-Efraim S, Aronson M, Bichowsky-Slomnicki L:
New antigenic component of Pasteurella pestis formed under specified conditions of pH and temperature.
J Bacteriol.
1961; 81(5): 704–14. PubMed Abstract
| Free Full Text
- 117.
Bichowsky-Slomnicki L, Ben-Efraim S:
Biological activities in extracts of Pasteurella pestis and their relation to the "pH 6 antigen".
J Bacteriol.
1963; 86(1): 101–11. PubMed Abstract
| Free Full Text
- 118.
Lindler LE, Klempner MS, Straley SC:
Yersinia pestis pH 6 antigen: genetic, biochemical, and virulence characterization of a protein involved in the pathogenesis of bubonic plague.
Infect Immun.
1990; 58(8): 2569–77. PubMed Abstract
| Free Full Text
- 119.
Yang Y, Isberg RR:
Transcriptional regulation of the Yersinia pseudotuberculosis pH6 antigen adhesin by two envelope-associated components.
Mol Microbiol.
1997; 24(3): 499–510. PubMed Abstract
| Publisher Full Text
- 120.
Lindler LE, Tall BD:
Yersinia pestis pH 6 antigen forms fimbriae and is induced by intracellular association with macrophages.
Mol Microbiol.
1993; 8(2): 311–24. PubMed Abstract
| Publisher Full Text
- 121.
Payne D, Tatham D, Williamson ED, et al.:
The pH 6 antigen of Yersinia pestis binds to beta1-linked galactosyl residues in glycosphingolipids.
Infect Immun.
1998; 66(9): 4545–8. PubMed Abstract
| Free Full Text
- 122.
Makoveichuk E, Cherepanov P, Lundberg S, et al.:
pH6 antigen of Yersinia pestis interacts with plasma lipoproteins and cell membranes.
J Lipid Res.
2003; 44(2): 320–30. PubMed Abstract
| Publisher Full Text
- 123.
Zav'yalov VP, Abramov VM, Cherepanov PG, et al.:
pH6 antigen (PsaA protein) of Yersinia pestis, a novel bacterial Fc-receptor.
FEMS Immunol Med Microbiol.
1996; 14(1): 53–7. PubMed Abstract
| Publisher Full Text
- 124.
Iriarte M, Vanooteghem JC, Delor I, et al.:
The Myf fibrillae of Yersinia enterocolitica.
Mol Microbiol.
1993; 9(3): 507–20. PubMed Abstract
| Publisher Full Text
- 125.
Beesley ED, Brubaker RR, Janssen WA, et al.:
Pesticins. 3. Expression of coagulase and mechanism of fibrinolysis.
J Bacteriol.
1967; 94(1): 19–26. PubMed Abstract
| Free Full Text
- 126.
Degen JL, Bugge TH, Goguen JD:
Fibrin and fibrinolysis in infection and host defense.
J Thromb Haemost.
2007; 5(Suppl 1): 24–31. PubMed Abstract
| Publisher Full Text
- 127.
Plow EF, Ploplis VA, Carmeliet P, et al.:
Plasminogen and cell migration in vivo.
Fibrinolysis Proteol.
1999; 13(2): 49–53. Publisher Full Text
- 128.
Sebbane F, Jarrett CO, Gardner D, et al.:
Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague.
Proc Natl Acad Sci U S A.
2006; 103(14): 5526–30. PubMed Abstract
| Publisher Full Text
| Free Full Text
| Faculty Opinions Recommendation
- 129.
Haiko J, Kukkonen M, Ravantti JJ, et al.:
The single substitution I259T, conserved in the plasminogen activator Pla of pandemic Yersinia pestis branches, enhances fibrinolytic activity.
J Bacteriol.
2009; 191(15): 4758–66. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 130.
Kienle Z, Emödy L, Svanborg C, et al.:
Adhesive properties conferred by the plasminogen activator of Yersinia pestis.
J Gen Microbiol.
1992; 138(Pt 8): 1679–87. PubMed Abstract
| Publisher Full Text
- 131.
Lähteenmäki K, Kukkonen M, Korhonen TK:
The Pla surface protease/adhesin of Yersinia pestis mediates bacterial invasion into human endothelial cells.
FEBS Lett.
2001; 504(1–2): 69–72. PubMed Abstract
| Publisher Full Text
- 132.
Hinnebusch BJ, Rudolph AE, Cherepanov P, et al.:
Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector.
Science.
2002; 296(5568): 733–5. PubMed Abstract
| Publisher Full Text
| Faculty Opinions Recommendation
- 133.
Wiechmann A:
Quorum sensing and the regulation of multicellular behaviour in Yersinia pseudotuberculosis.
(PhD Thesis. The University of Nottingham).
- 134.
Schraidt O, Lefebre MD, Brunner MJ, et al.:
Topology and organization of the Salmonella typhimurium type III secretion needle complex components.
PLoS Pathog.
2010; 6(4): e1000824. PubMed Abstract
| Publisher Full Text
| Free Full Text



Comments on this article Comments (0)