Keywords
Data display, Graphical user interfaces, Web service, SABIO-RK, kinetic parameters, SBML
This article is included in the Cytoscape gateway.
Data display, Graphical user interfaces, Web service, SABIO-RK, kinetic parameters, SBML
One of the main challenges for the modeling of biochemical systems is the availability of reliable information on the individual reaction steps and their kinetics from the literature. This information includes kinetic parameters with their rate equations as well as detailed descriptions of how these were determined1.
SABIO-RK (http://sabio.h-its.org/) is a manually curated database for kinetic data storing comprehensive information about biochemical reactions and their kinetic properties, with data manually extracted from the literature and directly submitted from lab experiments1,2. Available information comprises kinetic parameters with their corresponding rate equations, kinetic law and parameter types and experimental conditions under which the kinetic data were determined. In addition, information about the biochemical reactions and pathways including their reaction participants, cellular location and the catalyzing enzyme are recorded2.
The information in SABIO-RK is structured in datasets, so called database entries, which can be accessed either through the web-based user interface (http://sabiork.h-its.org/newSearch/index) or via web services (http://sabiork.h-its.org/sabioRestWebServices). Both interfaces support the export of the data in the Systems Biology Markup Language (SBML), a free and open interchange format for computer models of biological processes3.
Database entries are annotated with controlled ontologies and vocabularies based on Minimum Information Required for the Annotation of Models (MIRIAM4), e.g. reaction participants (e.g. small chemical compounds and proteins), as well as kinetic rate laws, and parameters. The annotation information is encoded in the form of RDF-based MIRIAM annotations, and additional XML based SABIO-RK specific annotations, e.g. for experimental conditions. These annotations integrate the kinetic information with external resources like ChEBI (https://www.ebi.ac.uk/chebi/)5, UniProtKB (http://www.uniprot.org/)6, PubMed (http://www.ncbi.nlm.nih.gov/pubmed/), or KEGG (http://www.kegg.jp/)7.
Despite the importance of kinetic information for computational modelling there has been no simple solution to visualize the database entries from SABIO-RK, and provide access to the network structure of the kinetic entries and the information encoded in the annotations.
In this work, we present cy3sabiork, an app for the visualization of kinetic data from SABIO-RK for Cytoscape, an open source software platform for network visualization8. cy3sabiork creates an intuitive visualization of kinetic entries in the form of a species-reaction-kinetics graph extended with kinetic information, which reflects the reaction-centered approach of SABIO-RK. Hereby, the kinetic information is accessible via a combination of graph structure and annotations of nodes, with provided information consisting of: (I) reaction details, enzyme and organism; (II) kinetic law, formula, parameters; (III) experimental conditions; (IV) publication; (V) additional annotations. cy3sabiork allows for easy comparison of kinetic data, visual inspection of the elements involved in the kinetic record and simple access to the annotation information of the kinetic record.
cy3sabiork was written in Java as an OSGi bundle for Cytoscape 3 using the app infrastructure. The bundle activator adds the cy3sabiork Action to the Cytoscape icon bar, which provides access to the JavaFX based cy3sabiork dialog. The cy3sabiork GUI is a combination of web based components handled in a WebView and classical GUI components. The combination of Swing and JavaFX is implemented based on a JFXPanel with JavaFX GUI updates in Platform.runLater, Swing GUI updates via SwingUtilities.invokeLater. The JavaFX approach allows for the integration of rich Web-based content using HTML/Javascript/CSS with Cytoscape. The GUI was created utilizing FXML based GUI definitions created in JavaFX Scene Builder, a JavaFX tool which allows to quickly design JavaFX application user interfaces by dragging UI components into a content view area. FXML code for the UI layout is created by the tool which was styled using CSS. The downside of the JavaFX-Swing hybrid approach are additional complexity in handling Events and EventListeners in different threads, some issues with Windows Management and Focus Handling, and full support of JavaFX in old felix OSGI containers.
SABIO-RK entries are retrieved via the web services using the RESTful API. SBML from the web service calls or the web interface export is imported using a Cytoscape Task created by the LoadNetworkFileTaskFactory. CyNetworks and CyNetworkViews for the imported kinetic entries are created by a CyNetworkReader registered for SBML files provided by cy3sbml9. During the app development the SBML CyNetworkReader was extended to support the SABIO-RK specific annotations and data. RDF based annotations are read with JSBML10,11 and hyperlinks to the respective resources are created by parsing the resources.
By implementing cy3sabiork as a desktop app, in comparison to a solely web-based solution with Cytoscape.js, tight integration with cy3sbml was possible, thereby providing rich functionality for the kinetic entries in SBML, like for instance access to the annotation information or the raw SBML files of the kinetic entries.
An overview over the typical cy3sabiork workflow is depicted in Figure 1. The main steps of operation are searching entries in SABIO-RK, loading entries in cy3sabiork, and visual exploration of results:
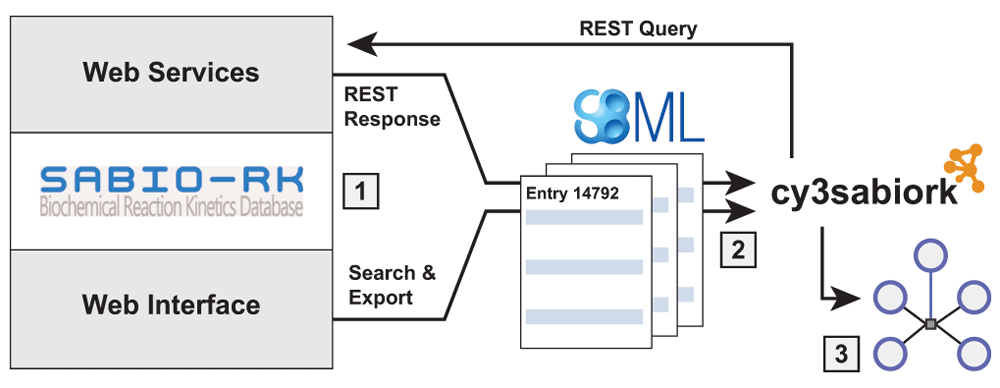
The main steps of operation are: (1) Searching entries in SABIO-RK: Kinetic entries are retrieved from SABIO-RK via calls to the RESTful web services or via searching the web interface and exporting selected entries as SBML; (2) Loading entries in cy3sabiork: The exchange format between SABIO-RK and cy3sabiork is SBML; (3) Visual exploration of results. The kinetic graphs for the imported entries are generated providing simple access to kinetic information and annotations. An example query with resulting SABIO-RK information and subsequent visualization is shown in Figure 2.
Searching kinetic entries. Kinetic entries can either be searched via the web services available from the cy3sabiork panel or directly in the SABIO-RK web interface available at http://sabiork.h-its.org/newSearch/index.
The web interface enables the search for reactions and their kinetics by either a free text search or an advanced search, which supports the creation of complex queries by specifying reactions by their participants (substrates, products, inhibitors, activators etc.) or identifiers (KEGG or SABIO-RK reaction identifiers and KEGG, SABIO-RK, ChEBI or PubChem compound identifiers), pathways, enzymes, UniProt identifiers, organisms (NCBI taxonomy12), tissues or cellular locations (BRENDA tissue ontology (BTO)13), kinetic parameters, environmental conditions or literature sources1,2. After finalizing the search the selected kinetic entries are exported as SBML.
The web interface supports the same query options, which can be added in the REST query GUI, but allows a direct import of the SBML without the requirement for search in the web interface and export and subsequent import of the SBML.
Example queries are depicted in Figure 2 and Figure 4 with resulting SBML available as Supplementary File S1 and Supplementary File S2.
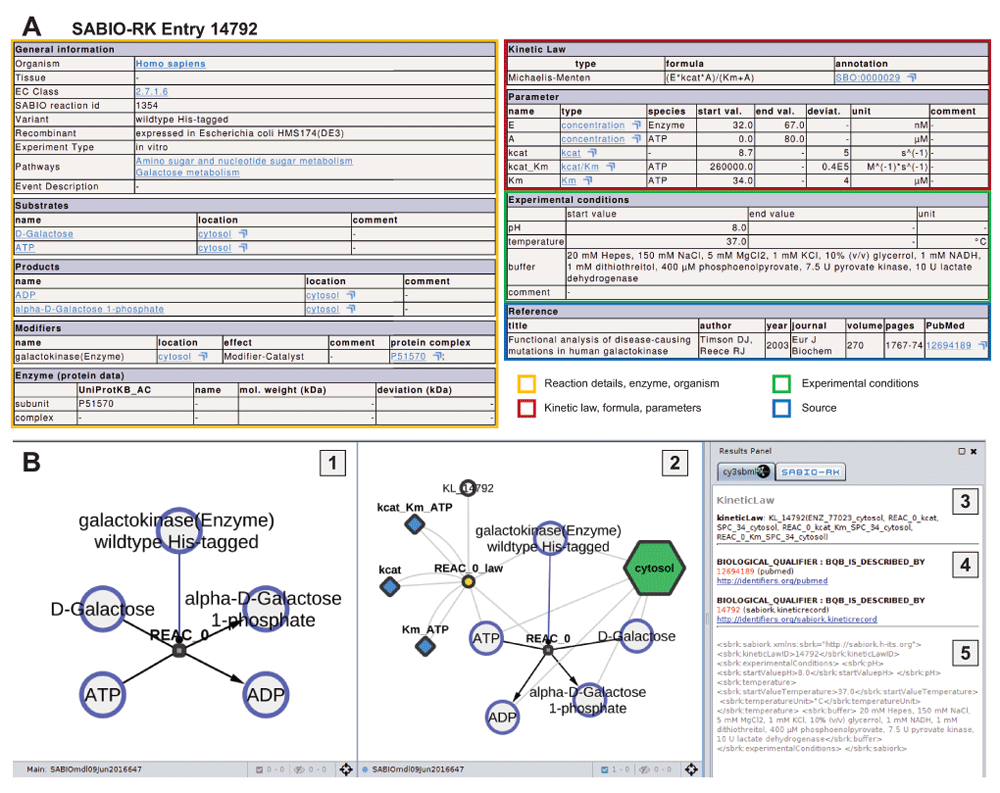
The kinetic entry 14792 for galactokinase (EC:2.7.1.6, UniProtKB:P51570) was retrieved via the web service query http://sabiork.h-its.org/sabioRestWebServices/kineticLaws/14792 (status 10-06-2016, SBML of query in Supplementary File S1). (A) Overview of kinetic information for SABIO-RK entry (http://sabiork.h-its.org/kineticLawEntry.jsp?viewData=true&kinlawid=14792) with color coding according to 1. (B) cy3sabiork information for entry 14792: (1) Resulting species-reaction-modifier graph. The galactokinase enzyme catalyzes the conversion of D-Galactose + ATP → α-D-Galactose 1-phosphate + ADP (see also Substrates, Products and Modifiers in A); (2) Kinetic graph with additional nodes for kinetic law, parameters and localization; (3) Selecting nodes in the graphs provides access to the annotation information and links to databases. In the example the kinetic law information is displayed in the Results Panel. (4) MIRIAM annotations with respective links to databases are available via the Results Panel; (5) Additional SABIO-RK annotations in XML for the experimental conditions are displayed in this section.
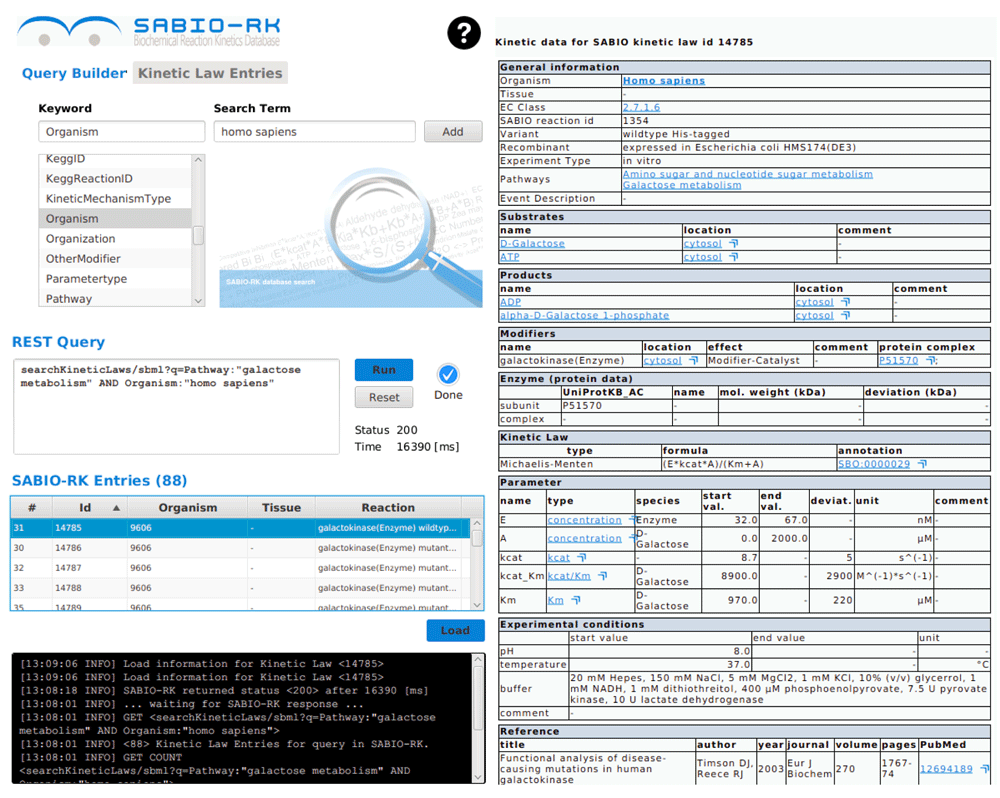
For human galactose metabolism 88 entries are available (status 28-06-2016, SBML of query in Supplementary File S2). For the selected Entry 14785 detailed information is provided on the right side.
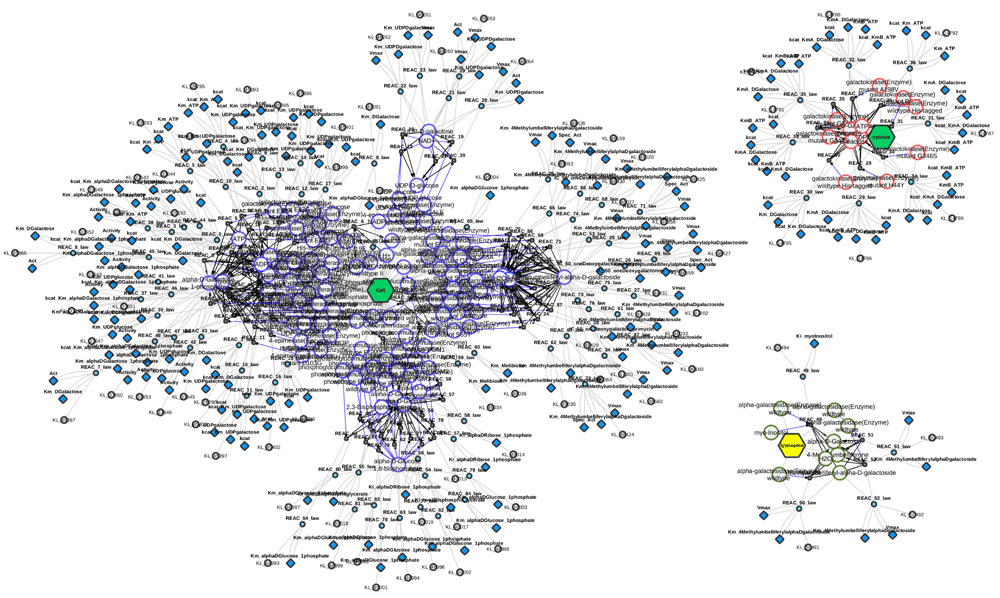
Graph of SABIO-RK kinetic information available for human galactose metabolism consisting of 88 entries (status 28-06-2016, SBML of query in Supplementary File S2). The kinetic graph consists of three clusters, separated based on the reported localization of the catalyzing enzyme. The lysosomal entries are non-canonical reactions in the galactose metabolism.
Loading kinetic entries. The SBML exported from web interface searches is imported as in Cytoscape using cy3sbml9 (File → Import → Network → File). For queries to the web services the response SBML is imported automatically without the need for additional file operations.
Visual exploration. The final step is the exploration of the kinetic entries in the species-reaction-modifier and the kinetic graph (e.g. in Figure 2 and Figure 4). The information from a wide range of resources and databases is integrated with the graph visualization of the kinetic records accessible as hyperlinks from the cy3sbml panel. Examples are the access to the source publication on PubMed from which the kinetic information was retrieved, the UniProtKB protein for which the kinetic information was measured, KEGG and ChEBI information for species involved in the reaction, or links back to the SABIO-RK database entry and reaction.
We applied cy3sabiork for kinetic parameter search and model construction of a kinetic model of galactose metabolism of the human liver (https://github.com/matthiaskoenig/multiscale-galactose) within the Virtual Liver Network (VLN) and Systems Medicine of the Liver (LiSyM) projects. A crucial step in building kinetic models of metabolism is the collection of kinetic information from the literature for the parameterization of the biochemical reactions. The search and visual exploration with cy3sabiork provides easy-access to a high-quality starting set of kinetic parameters and corpus of relevant publications for the processes of interest. Hereby, subsequent literature search and retrieval of referenced publications is simplified.
A representative SABIO-RK query for galactokinase (EC:2.7.1.6, UniProtKB:P51570), the first step of hepatic galactose metabolization is depicted in Figure 2 retrieving the SABIO-RK kinetic record 14792. A more complex query used for parameter search is depicted in Figure 3 and Figure 4 retrieving all kinetic records available for human galactose metabolism.
During model building publications with kinetic information not yet available in SABIO-RK were included in the database by the SABIO-RK curation service.
cy3sabiork is a Cytoscape app for visualizing kinetic data from SABIO-RK providing the means for visual analysis of kinetic entries from SABIO-RK within their reaction context. Herby, the integration of kinetic parameters with computational models is supported. The availability of direct links to annotated resources from within the network context of the kinetic records provides important information for the knowledge integration with computational models.
Software available from: http://apps.cytoscape.org/apps/cy3sabiork
Latest source code: https://github.com/matthiaskoenig/cy3sabiork
Archive source code as at the time of publication: http://dx.doi.org/10.5281/zenodo.5742814
License: GNU General Public License, version 3 (GPL-3.0): https://opensource.org/licenses/GPL-3.0
MK developed the app and wrote the manuscript. All authors were involved in the revision of the draft manuscript and have agreed to the final content.
This work was supported by the Federal Ministry of Education and Research (BMBF, Germany) within the research network Systems Medicine of the Liver (LiSyM, grant number 031L0054) and the Virtual Liver Network (VLN, grant number 0315741).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank the SABIO-RK team, the SBML community, and Cytoscape community for their support and help. A special thanks to the curation team of SABIO-RK including additional kinetic data in the database based on provided publications.
Supplementary File S1: Kinetic entry 14792.
SBML for query http://sabiork.h-its.org/sabioRestWebServices/kineticLaws/14792
Supplementary File S2: Kinetic entries for human galactose metabolism.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: I am the chief architect and project manager for Cytoscape, and am interested in seeing high quality apps added to the Cytoscape app store and made available to Cytoscape users.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 18 Jul 16 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (1)