Keywords
forensic science, flow cytometry, epidermal cell, touch DNA, autofluorescence, mixture
forensic science, flow cytometry, epidermal cell, touch DNA, autofluorescence, mixture
The difficulties associated with interpreting complex DNA mixtures are well known in the forensic community, and are becoming more prevalent with the sharp increase in ‘touch’ or trace samples among forensic laboratories’ caseloads1. Differentiating cell populations from individual contributors in a biological mixture before DNA analysis is a potential way to overcome this issue. While strategies exist to selectively label cell populations from distinct contributors based on their immunochemistry and then physically isolate cells from the mixture prior to DNA profiling2–4, there is a dearth of studies demonstrating cell separation techniques on touch samples. This is likely due to the fact that cell populations in these samples mostly, if not entirely, consist of fully differentiated keratinocytes which have limited reactivity to common molecular probes used to target surface antigens5,6.
An alternative approach is to avoid the need for probe binding by harnessing the intrinsic fluorescence of compounds found in or on epidermal cells. Here we report on our analysis of autofluorescence in the red region of the spectrum (650–670nm) of epidermal cells collected from surfaces touched by seven different individuals across multiple days, and the implications this may have for processing complex biological mixtures in forensic casework.
Touch samples were collected from seven volunteers using the following protocol which was approved by the VCU-IRB (#HM20000454_CR). Volunteers rubbed a sterile polypropylene conical tube (P/N 229421; Celltreat Scientific) for five minutes using their entire hand (i.e., palm and fingers). Cells were collected from the surface with six sterile pre-wetted swabs (P/N 22037924; Fisher Scientific) followed by two dry swabs. To elute the cells into solution, the swabs were manually stirred then vortexed for 15 seconds in 10 mL of ultrapure water (18.2 MΩ∙cm). The entire solution was then passed through a 100 µm filter mesh prior to flow cytometry. Flow cytometry analysis of eluted cells was performed on the BD FACSCanto™ II Analyzer (Becton Dickinson) equipped with 488 nm and 633 nm lasers and a 660/20 nm detector filter. Channel voltages were set as follows: Forward Scatter (FSC, 150V), Side Scatter (SSC, 200V) and Allophycocyanin (APC, 250V). FSC and SSC channels were used to gate intact corneocytes for subsequent autofluorescence analysis. Gating of cell populations and generation of histogram profiles for each contributor was performed using FCS Express 4.0 Flow Research Edition (De Novo Software, Inc.).
Fluorescence histograms of individual cell populations from different donors are shown in Figure 1. For ease of comparison and visualization, profiles have been overlayed and grouped by the day on which cells were deposited, collected, and analyzed by flow cytometry. Clear differences in the red fluorescence (APC) channel are observed between several pairs of donor cell populations, particularly J16-D02 during the first experiment and J16-S07 in the second experiment (Figures 1a and 1b respectively; Table 1). Most experiments resulted in one or more contributor cell population(s) whose fluorescence profile(s) could be distinguished from the others collected that day, such that a fluorescence intensity gate could be designed that would be expected to capture that contributor’s cells to the exclusion of (or minimal contribution of) cells from other contributors. However, significant and/or complete overlap was observed between many donor pairs (e.g., A42-B17 in Figure 1a; I66-S07 in Figure 1d). Sometimes, overlap of fluorescence distributions was such that gating could potentially separate the contributors into two or more groups (e.g. Figure 1d: A42, B17, I66, R12 and S07 in one group; D02 and J16 in another group). All contributors from the final experiment exhibited overlapping fluorescence histograms (Figure 1e).
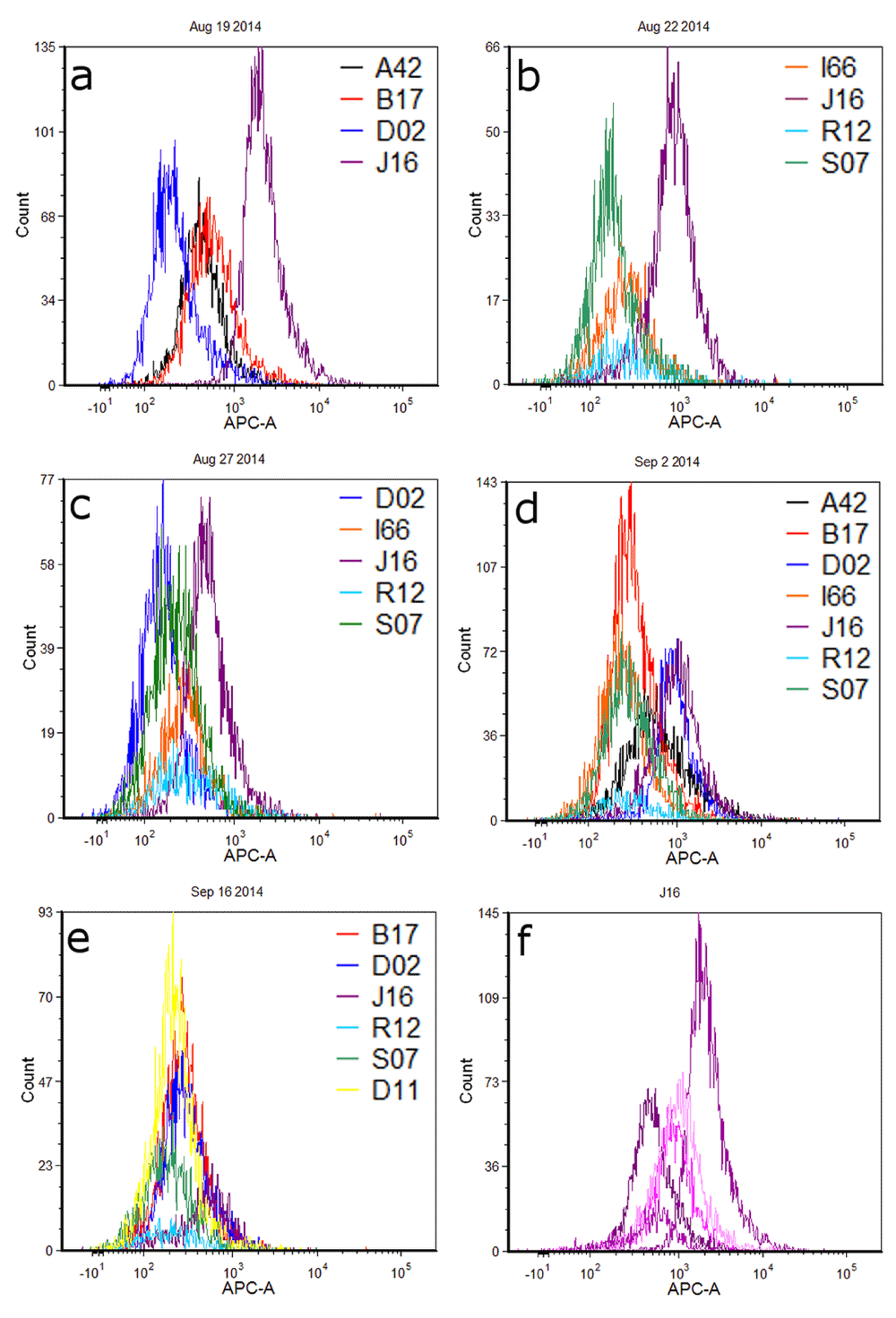
Panels a–e show different combinations of donors cell populations each sampled and analyzed on the same day. Figure 1f is a histogram overlay of cell populations from contributor J16 across five different experiments.
| Fig 1a | Fig 1b | |||||||
|---|---|---|---|---|---|---|---|---|
| Donor | Mean | Median | # Events2 | Donor | Mean | Median | # Events | |
| A42 | 540 | 427 | 3903 | I66 | 341 | 253 | 1573 | |
| B17 | 743 | 556 | 4625 | J16 | 996 | 842 | 3375 | |
| D02 | 305 | 212 | 5158 | R12 | 497 | 252 | 599 | |
| J16 | 2606 | 2024 | 6475 | S07 | 236 | 177 | 2497 | |
| Fig 1c | Fig 1d | |||||||
| Donor | Mean | Median | # Events | Donor | Mean | Median | # Events | |
| D02 | 208 | 160 | 3653 | A42 | 959 | 554 | 4320 | |
| I66 | 372 | 276 | 1983 | B17 | 409 | 307 | 7727 | |
| J16 | 635 | 491 | 3767 | D02 | 1114 | 907 | 3524 | |
| R12 | 469 | 298 | 1090 | I66 | 314 | 244 | 5014 | |
| S07 | 279 | 226 | 3751 | J16 | 1245 | 982 | 4702 | |
| R12 | 457 | 260 | 861 | |||||
| S07 | 376 | 277 | 4676 | |||||
| Fig 1e | Fig 1f | |||||||
| Donor | Mean | Median | # Events | Donor | Mean | Median | # Events | |
| B17 | 349 | 280 | 3665 | J16a | 2606 | 2024 | 6475 | |
| D02 | 362 | 287 | 3041 | J16b | 635 | 491 | 3767 | |
| J16 | 589 | 515 | 1156 | J16c | 589 | 515 | 1156 | |
| R12 | 302 | 208 | 493 | J16d | 996 | 842 | 3375 | |
| S07 | 259 | 190 | 2028 | J16e | 1245 | 982 | 4702 | |
| D11 | 276 | 220 | 4230 |
1Data is organized according to the histogram overlays shown in Figure 1. Mean (arithmetic) and median values are in relative fluorescent units (RFUs).
Cell populations from J16 and D02 showed a great deal of disparity in fluorescence intensity in the first experiment, such that overlap between these populations was minimal (Figure 1a). There was somewhat less distinction – and thus more overlap – observed between the same contributors during a second replicate (Figure 1c); during a third, overlap between the two populations was substantial (Figure 1d). As these results suggest, fluorescence intensity values for cell populations derived from any given contributor varied in distribution across replicate experiments on different days. Figure 1f shows overlayed histograms for J16 cell populations; mean fluorescence intensity values ranged from 589 to 2606 relative fluorescence units (RFUs) across five sampling days (Table 1).
The underlying cause of red autofluorescence in these epidermal cell samples is currently unclear. Cells deposited through touch are likely primarily derived from the outermost epidermal layer (stratum corneum) which can contain a number of fluorescent compounds including tryptophan and tyrosine7,8, melanin, keratins, NADH and flavins9, lipofuscins10, and porphyrins and porphyrin precursors11,12. However, many of the corresponding emission maxima for these molecules occur at shorter wavelengths than what was examined in this study (e.g., amino acids, keratin, NADH, all have maxima below 550nm9). Porphyrin molecules exhibit emission maxima between 630–680nm11. Their abundance within the epidermis may be influenced by bacteria on the skin that produce porphyrin molecules with similar fluorescence emission profiles13. Exogenous sources such as plasticides14 or other biological compounds (e.g., chlorophyll15) may also produce fluorescence, and could potentially be transferred to donors’ hands and subsequently to the tube surface (with cells) through touch or contact.
Regardless of the ultimate source for the observed differences in cell population fluorescence, this initial data set indicates that autofluorescence may be a useful marker for distinguishing between cell populations in a mixture. The non-destructive nature of flow analysis and the fact that autofluorescence monitoring does not require special reagents beyond those maintained in any laboratory (e.g. no probes required) are advantages when considering their potential front-end use in forensic analyses.
The variation across multiple samples from the same donor suggests that the level of autofluorescence is likely not a unique or identifying feature for a particular individual. However, to be of use in separating components of a biological mixture, a feature need not be unique; it simply needs to be distinctive among the contributors to that particular mixture. The ability to separate out even one contributor (or to separate a mixture of four contributors into two mixtures of two) may render the remaining mixture more interpretable in downstream DNA analysis. Further, the possibility that some combination of endogenous and/or exogenous factors could impart distinct optical properties to contributor cell populations in a particular mixture sample warrants further exploration.
Future efforts will continue to focus on isolating the molecule(s) responsible for fluorescent differences in touch epidermal cells through a combination of targeted immunofluorescent assays, chemical characterizations, and complex spectral analysis of autofluorescent profiles. Additionally, we are working on using optical signatures such as these to facilitate physical isolation of epidermal cell populations using flow cytometry-based strategies such as fluorescent activated cell sorting (FACS) for the purposes of generating single source genetic profiles from touch mixtures. Although previous work suggests that analyzing DNA profiles directly from isolated epidermal cells may be a challenge due to the prevalence of extracellular or ‘cell-free’ DNA in touch samples16, the sheer quantity of cells that may be recovered from these sample types (up to ~1×105,16) may help to overcome such obstacles.
F1000Research: Dataset 1. Flow cytometry source data for individual contributors, 10.5256/f1000research.8036.d11374917
CE conceived the study. CE, CS, KP, EB, and YK designed the experiments. CS, EB, YK carried out the research. KP assisted with data analysis and provided expertise in the area of forensic casework. CE, CS, and KP prepared the first draft of the manuscript. All authors were involved in the revision of the draft manuscript and have agreed to the final content.
This project was funded by the National Institute of Justice Award number 2013-DN-BX-K033 (PI: Ehrhardt). Flow cytometry services in support of the project were provided by the VCU Massey Cancer Center, supported in part with funding from NIH-NCI P30CA016059.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors gratefully acknowledge Daniel Conrad and Julie Farnsworth for providing technical assistance for this project.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
References
1. Perlin MW, Legler MM, Spencer CE, Smith JL, et al.: Validating TrueAllele® DNA mixture interpretation.J Forensic Sci. 2011; 56 (6): 1430-47 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
References
1. Quinones I, Daniel B: Cell free DNA as a component of forensic evidence recovered from touched surfaces.Forensic Sci Int Genet. 2012; 6 (1): 26-30 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 16 Feb 16 |
read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)