Keywords
CRM, LIMS, Data collection, Data-warehouse, Integration Software-as-a-service (SaaS)
CRM, LIMS, Data collection, Data-warehouse, Integration Software-as-a-service (SaaS)
The nature of modern research is to collect ever larger and ever more complex data sets in order to address present day scientific problems, which in turn requires more sophisticated data management1. This increase in size and complexity is particularly apparent in the biomedical research domain, where software tools are having to be rapidly developed to meet these data challenges. This software development is often driven by large research groups who have the resource and expertise to meet their needs. This inevitably results in highly customised software solutions and infrastructure that may not be reusable elsewhere.
Smaller research groups often do not have the resources or expertise to do the equivalent software development themselves. They are then left with no other option than to buy off the shelf (often proprietary) tools in order to meet their needs, or to use tools not designed to do research. Proprietary software tools are often expensive, and often do not allow any user customisation, hindering further reuse. This can then lead to research groups being charged further to have the software modified to meet their requirements.
An increasingly viable option is to use open source software to build the required research platform. This is an approach that is being actively pushed at ever higher levels - the UK government actively encourages the use of open source software, and have had policies mandating its use when there is no significant cost difference due to its added flexibility (https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/78959/All_About_Open_Source_v2_0.pdf).
Increasingly applications are moving away from being installed locally on client machines (e.g. desktops, laptops etc.) and are being accessed via web browsers e.g. email, word processing, file storage, etc. This has the effect of making them easier to maintain since there is a central install of the software, instead of many local installs on a variety of hardware/operating system/software combinations. It also means that data is not stored locally, thus reducing the risk of data loss or disclosure by the client machine (although there are other risks associated with centralised systems). This approach is often referred to as software as a service (SaaS).
Here we outline the technical aspect of the Biomedical Research Infrastructure Software as a Service Kit (BRISSKit) (http://www.brisskit.le.ac.uk/) project that builds on the trend for open-source and online applications. A subsequent paper (Jonathan A. Tedds, Neil Beagrie, Shajid Issa, Oliver W. Butters, Josh Vande Hey, Scott Wilson, Rebecca C. Wilson, Rowan Wilson, Andrew Charlesworth, and Paul R. Burton - Unpublished report, 2016) will describe use case implementations, the underlying business case, sustainability options, service vision for the platform and proposed further developments and applications.
There are various web based and open source applications being used (and developed) in biomedical research, for example: the Galaxy project (http://galaxyproject.org) which focusses on genetics analysis, Harvest (http://harvest.research.chop.edu)2 which is a biomedical data discovery framework, ARIES Explorer (http://www.ariesepigenomics.org.uk) which is an epigenetic browser, and tranSMART (http://transmartfoundation.org)3 which is a translational biomedical research knowledge management platform.
There have been comparable projects in other disciplines, the Virtual Observatory (http://ivoa.net), as developed in the AstroGrid project, e.g. 4 in particular, has influenced the development of this project.
Here we focus on four different open source applications that are applicable to biomedical research studies: CiviCRM, OpenSpecimen, Onyx and i2b2. These four applications were chosen as they formed the core part of the National Institute for Health Research Leicester Cardiovascular Biomedical Research Unit (NIHR-LCBRU) informatics platform, which has been used to recruit thousands of participants into research studies in the East Midlands in the UK (http://www2.le.ac.uk/research/current-research/bru/our-research/research-facilities/informatics-platform).
CiviCRM. CiviCRM (https://civicrm.org) is a web-based open source (GNU AGPL v3) constituent relationship management tool built on top of Drupal (https://drupal.org). It is designed to manage the contact details of individuals and their relationships with things. It also manages the means to contact individuals. It can be configured to have almost any number and type of data field for each individual added to it. This configurability makes it an ideal tool to track study participant details, e.g. names, addresses, phone numbers etc. CiviCRM then adds the ability to model relationships between participants e.g. mother-child, doctor-patient, work colleague etc. CiviCRM also adds the concept of organisations, these can be used to model e.g. a household which a number of participants could belong to, a hospital that participants are patients in etc. Furthermore, CiviCRM adds extra value in its case functionality - this allows a series of activities to be defined which may model individual stages of a biomedical study, for example: fill in a consent form, take a blood sample, schedule an appointment etc. These activities may be linked together sequentially, or in a more complex non-sequential way with additional conditional logic. This mirrors the work flow that a biomedical research study may have.
Given these features it is easy to see how CiviCRM could serve as the main study management application in a biomedical study.
CiviCRM is written in PHP, and uses a MySQL database. The source code is available at: http://sourceforge.net/projects/civicrm
OpenSpecimen. OpenSpecimen (formally known as CaTissue) (http://www.openspecimen.org/) is an open source (BSD 3-clause licence) web-based biobanking management system. It was originally developed as part of the U.S. National Cancer Institute’s caBIG program as CaTissue. CaTissue was then forked by a commercial company and re-branded as OpenSpecimen (still keeping it open source). They now maintain the core code base, and offer support and hosting. It has a highly configurable object model, making it possible to model almost any type of storage infrastructure used in biobanking (e.g. boxes on shelves, in freezers, in rooms, in buildings etc.). Samples can then be put into the system and tracked as e.g. containers are moved or samples are checked out etc.
OpenSpecimen is written in Java and uses a MySQL database. The source code is available at: https://github.com/krishagni/openspecimen
Onyx. Onyx is an open source (GNU GPL v3) web-based data collection tool developed by the Canadian company OBiBa (http://obiba.org). It is primarily used to collect questionnaire data from study participants, and was developed with the aim of collecting data from over 300,000 participants in the Canadian Partnership for Tomorrow program. Onyx is written in Java and uses a MySQL database. It is available at https://github.com/obiba/onyx
i2b2. i2b2 (Informatics for integrating biology and the bedside) (https://www.i2b2.org) is an open source (custom written licence - https://www.i2b2.org/software/i2b2_license.html) data warehouse framework built by Partners Healthcare System5. At its core are several ‘cells’, each providing a specific piece of functionality e.g. identity management, ontology management, data storage, natural language processing, web client etc. These cells are arranged together into the i2b2 ‘hive’, in which each cell can communicate with the others via XML based web services. This allows biomedical data from multiple sources to be stored with ontological codes and presented side by side. The end users can then query the multi-source data using a web browser. Using this functionality some analysis can be done on the data, or new cohorts of participants generated based on some phenotypic criteria.
i2b2 is written in Java and uses a Microsoft SQL server, an Oracle or a postgreSQL database. It is available at https://www.i2b2.org/software/
Even with the suitability and availability of the applications above, they have not been widely adopted in the field. We think this is due to the presence of three main barriers to widespread adoption: installation and maintenance, hosting and integration.
Installation and maintenance. Each of the user facing applications are freely available for anyone to download and install without any charge or mandatory contracts. They are all provided with some installation instructions which will guide a user through installing and configuring the applications. However, a high degree of technical expertise is still usually required to perform these steps.
Once the applications have been installed they need to be maintained, this will usually involve upgrading the software when new versions are released, troubleshooting any problems with the software, and regularly backing up the data. Again, a high degree of technical expertise is required for this.
Hosting. An important decision to take when planning on running one of these user applications is where it should be installed. Since each application is accessed through a web browser it is vital that consideration is given to accessibility and security. A key factor in the choice of host has to be the physical location of data centres, since each data centre is subject to the local laws of the country in which it is physically based. At a more local level, hosting providers do not all offer the same service, some have high levels of security standards which they evidence with certifications like ISO 27001, others do not. Some have a direct connection to the UK academic network JANET (http://ja.net), others to the UK NHS network. Wherever sensitive patient data is involved a careful and thorough approach to information governance is essential. In the UK compliance with the NHS Information Governance Toolkit (https://www.igt.hscic.gov.uk/) is often required.
Integration. Each application is a valuable resource in its own right, but even greater value can be achieved when they are integrated together. A simple example of this would be where multiple data collection tools are able to automatically export their data and have it imported into a central data warehouse. This would allow data from multiple sources to be analysed at once.
This integration process is perhaps the most difficult to overcome of the barriers to widespread adoption, since it requires detailed technical knowledge of multiple systems.
While the three main barriers to widespread adoption (the technical know-how to install and maintain the applications, the facility to host the applications in a secure environment, and the facility to integrate the applications together and to external applications) have been individually overcome, to a greater or lesser extent by various groups, a significant amount of development and time is generally needed.
It is with this backdrop that the BRISSKit project was conceived and exists - it aimed to provide access to a suite of mature open source applications, hosted in a secure environment, integrated together and accessed via a web browser. The intended end-user base for BRISSKit was that of groups with multiple users who may or may not be co-located, and who do not necessarily have the technical experience (or resources) to set up and maintain the software themselves.
The user facing applications chosen to achieve this are those outlined earlier i.e. CiviCRM (v4.1), OpenSpecimen (v1.2-plus2.0), Onyx (v1.9) and i2b2 (v1.5).
Although BRISSKit has been primarily focused at biomedical research groups, these tools can be adopted in a similar way by other disciplines also.
This section describes how BRISSKit addressed the three main barriers to widespread adoption outlined earlier. The core infrastructure design subsection covers installation, maintenance and hosting, and the subsequent section addresses integration.
Below is an outline of the main design choices and components of the core infrastructure, on which the client facing software is installed. It begins at the bottom of the stack with the virtualisation/operating system layer, it then moves up a layer and addresses the configuration of the operating system, then up to the actual install of the software, finally moving to the overarching monitoring.
Virtualisation layer. A key objective for the infrastructural design is to make the platform easily accessible, deployable and scalable. A cloud based environment facilitates these needs and allows the rapid provision of new resources as needed.
An early development decision was to use an Infrastructure as a Service (IaaS) provider. This allowed the maximum degree of customisation when designing and running the platform. With IaaS, virtual machines (VMs) can be provisioned with the required specifications as needed. In order to take into account the issues around the physical location of data centres, a UK based hosting provider with UK based data centres was used - Eduserv’s cloud compute solution (http://www.eduserv.org.uk). Eduserv also has a direct connection to JANET, allowing fast transfer of data to/from UK universities. Eduserv provided VMWare’s vCloud Director interface (http://www.vmware.com/products/vcloud-director/) giving a software defined data centre.
One of the features missing from the Eduserv offering was direct NHS connectivity. In order to meet this need we worked closely with the University Hospitals Leicester Trust to develop the BRISSKit platform to run on their internal (N3 connected) infrastructure, which ran VMWare’s vSphere (v5).
While both of the above infrastructures use proprietary VMWare software, none of the proprietary methods (e.g. for the provisioning of new VMs) were used i.e. the BRISSKit platform could be run on any virtualisation technology or provider (e.g. Amazon, Azure etc.).
Encapsulation. When designing a cloud based software solution it is important to consider how different users may interact with one another. Clearly each research group wants their data to be completely demarcated from any other research group. With this in mind the platform was designed to completely encapsulate one instance of the software stack, not allowing any communication with any other instances of the stack. Initially this was achieved using VMWare’s vApp functionality, with each application having its own VM. However, this was deemed to be too closely tied to one vendor’s proprietary methods. Later this was moved to a software defined vApp (still with a separate VM for each application). Through an automated method the VMs were grouped together and isolated by the firewall, so could only communicate with other VMs in the same vApp. This allowed a way of building an instance of the infrastructure in a way that is completely agnostic to the underlying virtualization technology. This can be visualised in figure 1 where two instances of the software stack are shown running side by side, but are isolated from one another.
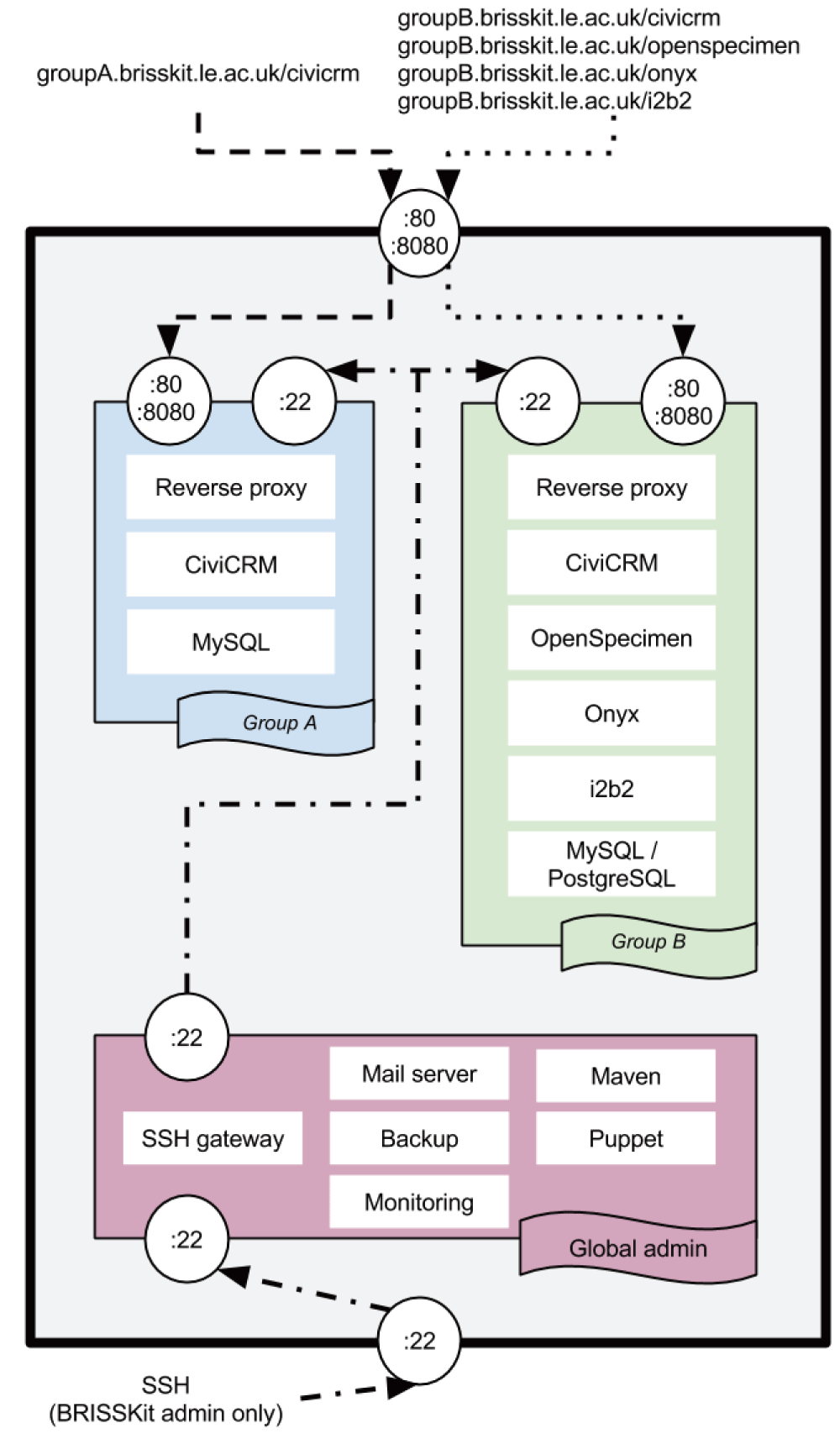
Two independent research groups are shown (group A and group B) illustrating the encapsulation of resources. The lines show the only network routes into and out of the infrastructure.
Configuration management. A key design choice for the management of the VMs was to use the Puppet configuration manager (v2.7) (https://puppetlabs.com/) to manage all software installation, configuration, user access etc. in the VMs, up to the point where the user applications can be installed. This allows the configuration to be managed in a declarative way, and information about VMs to be collected into a central resource. There are several benefits to this approach; the declarative nature means that the client VMs end up being configured in the required way- the process taken to get there is not important. Development of scripts which handle the configuration in a specific order is therefore not necessary. It also makes the type of guest operating system (OS) on the VM less important since e.g. software listed as being required is installed by the Puppet client, regardless of the different software management packages each OS has.
Another advantage of all configuration being managed centrally is that there is no real need to log onto specific client VMs to make changes such as new firewall rules etc. One final major advantage of using Puppet is the central gathering of information about the clients. Puppet has, at its core, a central database which logs information about the state of the clients, this can include server info such as uptime etc., as well as versions of software installed, which is a useful tool for auditing.
All of the configuration is put together in a central catalogue that each VM can query. Based on the role of the VM the catalogue items are automatically configured to e.g. set up the appropriate firewall rules etc. The overarching catalogue items developed as part of this project are listed in table 1.
Instantiation. Encapsulation is achieved based on the name and role the VM is given when it is created. As an example, imagine a research group using BRISSKit called groupA, a VM called groupA-civicrm would be created. Puppet looks at this name and deduces that it belongs to the software defined vApp for groupA, and it’s role is as a CiviCRM VM. Puppet then takes this new VM’s IP address and allows access through the firewalls on each of the other VMs in this vApp to this new VM, and adds a hostname entry to each. The other VMs in this vApp will then be able to connect to groupA-civicrm as required. At the same time the reverse proxy has an entry added to its rules so any web traffic meant for CiviCRM is directed to the correct VM. It also automatically adds an entry to the central nagios monitoring server. Once this generic configuration management has finished, Puppet then applies the catalogue entry relevant for this role (in this case - installing Apache, PHP libraries and the MySQL client). The end point of this process is a VM which is ready to have the client software (CiviCRM in this case) installed on it.
Web access. As mentioned previously, all of the user applications are primarily accessed via a web interface. To tie these together a reverse proxy was implemented, this allowed a subdomain to be defined per research group and each application to sit below that, e.g.
• groupA.brisskit.le.ac.uk/civicrm
• groupA.brisskit.le.ac.uk/openspecimen
• groupA.brisskit.le.ac.uk/i2b2
• groupB.brisskit.le.ac.uk/civicrm
• etc.
The open source reverse proxy software Pound (v2.5) (www.apsis.ch/pound) was used to achieve this.
All web traffic was encrypted from the client to the reverse proxy with SSL certificates.
Client software installation and maintenance. A significant amount of effort was devoted to automating the install of each user facing application. This gave the benefit of being able to deploy an application very quickly and in a standard way, thus avoiding any mistakes that may occur due to human error. In order to do this a common platform was defined in which to start the installation from. This consisted of an 64bit Ubuntu 12.04 Long Term Support operating system (http://www.ubuntu.com) with all configuration centrally managed with Puppet. This meant that the installation process for each application consisted of deploying a new instance of Ubuntu, the Puppet master would then automatically configure it ready to run the application installation.
The application installation scripts were all managed in version control, and then built with the software build tool Maven (v2.2) (http://maven.apache.org). Periodically, copies of the Maven built artefacts were deposited in our local Nexus repository, along with all the relevant dependencies, or links to remote repositories (Nexus software: http://www.sonatype.com/nexus-repository-oss). This process gave a standard installation procedure that could be followed for each of the applications, despite them requiring very different processes and dependencies in their native form.
Extra guidance and documentation on the install of the core applications that has been built up over the project is also available on the project website at http://www.brisskit.le.ac.uk.
Reporting. An essential part of any infrastructure is monitoring of servers and applications. Within BRISSKit, Nagios (v3) was implemented (https://www.nagios.org/). Nagios is an open source (GNU GPL v2) monitoring solution that follows the client-server model. All of the VMs report in periodically on their status to the central Nagios server. Different VMs report different measures based on their role. There are a core set of measures (CPU load, disk usage etc.) that all report, but on top of this there are others - the MySQL VM reports the status of its MySQL server for instance.
This set up facilitated the proactive monitoring of the infrastructure and fixing of problems as they happened, moreover, developing problems could be fixed before they manifested. This also served as a means of measuring resource usage and therefore facilitating cost effective use of compute resource.
The distributed nature of the BRISSKit infrastructure meant that the Nagios server could not always instigate active checks. Passive checks were therefore run across the infrastructure, instigated from the clients. This was achieved by using the NRDP Nagios plug-in (https://exchange.nagios.org/directory/Addons/Passive-Checks/NRDP–2D-Nagios-Remote-Data-Processor/details) on the clients along with scheduled cron jobs. All of which was managed with Puppet.
Use case. In order to describe the integration model developed, an end to end use case needs to be outlined first. Assume a study is using all four applications, the study behaviour can be defined in CiviCRM, into which study participants can be added. The study definition then means that these participants could be passed to the other applications in the stack. Onyx would then be ready for a participant to fill in a questionnaire as it would have e.g. a name pre-populated and an appointment time specified. OpenSpecimen would also be ready for a sample to be input. Once the data is collected it is automatically imported into i2b2, and data from the different sources (i.e. Onxy and OpenSpecimen) about the same individual is joined together. It is here that analysis of the data would happen. If there was a new cohort that emerged from the analysis, then they could be imported back into CiviCRM to be re-identified, and then invited back for more tests.
It is with this use case in mind that we describe the integration.
The layers. The BRISSKit architecture can be thought of as different layers, each containing a different category of application. Each layer can communicate with the other layers in a well defined way via the internal application programming interface (API). In this way the architecture can be split into three distinct layers based on the category of application it contains - management, data collection and data warehousing. This allows the user facing applications to be categorised into one of those three; CiviCRM - management, OpenSpecimen and Onyx - data collection, and i2b2 - data warehousing. The layers are stacked, with the management layer at the top, the data collection layer in the middle, and the data warehousing layer at the bottom - see figure 2. Messages are sent between the layers, through the internal API.
This layered architecture also serves as a means to categorise user access. For example, administrative staff may have access to identifiable contact details in CiviCRM, lab and data collection staff may have access to OpenSpecimen and Onyx and researchers access to non-identifiable data in i2b2. In this way data is very clearly segregated based on user roles.
The internal API. The layered architecture allows us to define standard messages that get sent between the layers. Each individual application must then adhere to the standard message definition when it sends messages between layers. Figure 2 illustrates this.
By adopting this layered structure it becomes relatively straightforward to add new components to the software stack - they just need to be able to communicate with the adjacent layers. This can be achieved by wrapping each new application up in such a way that it only communicates via the wrapper (the orange part in figure 2).
In order to make the API easy to use it is being developed as a REST interface. This also allows the API to be portable and scalable. Table 2 lists some pseudo-API calls to illustrate the layers.
The layers correspond to data going from the management layer (M) to the data collection layer (C), and to the warehousing layer (W).
The approach taken to implement the internal API will be different for each application due to the different technologies used in each.
CiviCRM has a mature REST API allowing interaction with it’s core functions. In order to achieve the BRISSKit internal API calls the native CiviCRM API was wrapped in our own functions. The add_participant_to_(object) function was triggered when participants were added to a study in CiviCRM and had an appointment booked. This passed the relevant participant information to the data collection tools.
OpenSpecimen did not have a mature API available during the development of this phase of the project (it does now). In order to fit it into our infrastructure we therefore had to call core Java classes to manipulate the data. The developed integrations accepted a participant from CiviCRM and created the sample collection stub, then once the samples were taken the relevant information extracted and pushed to i2b2. Now the API is more mature (and RESTful) these calls could be migrated to use it.
Onyx does not have an API available. It does however have routines that can be called to load and export data. These were wrapped into BRISSKit functions to allow participants to be added, and for the data to be extracted and pushed to i2b2.
i2b2 does not have an API as such, it’s modular design uses internal API calls to facilitate communication between cells, but this is not intended for external use. Modifying the Clinical Research Chart (CRC) loader facilitated loading data from the collection tools (import_data). We also developed a package so that groups of participants defined in i2b2 could be pushed back to CiviCRM. This enabled a work flow where a researcher could define a group of participants in i2b2 based on questionnaire answers and availability of samples without having access to their contact details, push that group back to CiviCRM to be re-identified (reidentify_cohort), and then followed up for more tests.
ID numbers and linking data. CiviCRM generates a unique random ID for each participant. This is the pseudonymised ID pushed to each data collection application in the layer below. Once the data is exported from each data collection application it is linked together at the data warehousing layer using this ID.
Ontology builder. Ontologies are an important aspect of the BRISSKit data warehousing layer since they form an integral part of i2b2’s functionality, and give the data meaning. Part of the integration of new applications in the stack therefore requires i2b2 to have an ontology describing the data in order to be able to understand it. Within the infrastructure, OpenSpecimen and Onyx automatically generate their own ontologies based on the data structures in the applications. This gets passed to i2b2 so the data can be queried.
i2b2 does not specify which ontology it has to use, so the automatically generated ones are considered ‘nominal ontologies’, i.e. they do not necessarily conform to one of the standard ontologies such as e.g. SNOMED CT.
Since it is possible that not all data will come from the core applications (i.e. CiviCRM, Onyx and OpenSpecimen) - some may need to be imported into i2b2 from external sources - an ontology building tool was developed. This plugged into the National Center for Biomedical Ontology’s BioPortal service (http://bioportal.bioontology.org/) and allowed ontology codes from standard ontologies to be used.
Given the modular nature of the infrastructure design, and the relative ease of integrating new applications, we are planning on adding more applications to the stack. Opal (http://obiba.org/node/63) is a data warehouse built by the same group that developed Onyx. One of the key features of Opal is that it integrates with the analysis tool DataSHIELD6,7, allowing analysis across multiple data warehouses in a non-disclosive way. Early implementations focussed on simultaneous epidemiological analysis across multiple international birth cohorts but this approach could be just as valuable in e.g. enabling analysis of studies across multiple hospital sites in one territory. At the data collection layer we have done some work to integrate the Research Electronic Data Capture tool, REDCap (http://www.project-redcap.org), into the stack. Going forward we will develop this integration so it becomes a core part of the BRISSKit stack, and is available to groups who have the appropriate agreement to use it. The internal API needs to be formalised somewhat, then an external API can be developed to connect to it, thus making it easy to integrate with third-party systems e.g. importing data from external systems. It is likely that we would use openESB (http://www.open-esb.net/) to further extend this functionality.
We have shown that it is possible to install and integrate a suite of mature open source applications for use by biomedical researchers. Moreover we have demonstrated that these applications can be installed in a cloud environment and isolated in such a way that multiple research groups can share the same infrastructure, but have their data completely separate from one another. We believe this a viable alternative to local installations of proprietary software, and logically leads to the idea of a research platform as a service that could be offered to research groups. A subsequent paper (Jonathan A. Tedds, Neil Beagrie, Shajid Issa, Oliver W. Butters, Josh Vande Hey, Scott Wilson, Rebecca C. Wilson, Rowan Wilson, Andrew Charlesworth, and Paul R. Burton - Unpublished report, 2016) will describe use cases and outline sustainability options for the BRISSKit platform.
Latest source code: https://github.com/brisskit-uol
Archived source code as at time of publication:
CiviCRM http://dx.doi.org/10.5281/zenodo.573848
Onyx install: http://dx.doi.org/10.5281/zenodo.573809
OpenSpecimen install: http://dx.doi.org/10.5281/zenodo.5737710
i2b2 install: http://dx.doi.org/10.5281/zenodo.5737411
Puppet: http://dx.doi.org/10.5281/zenodo.5736512
Software license: BSD 3-clause license.
OB, SI, JL, RP & RF all contributed to the overall technical development of BRISSKit. TB designed the ontology builder aspect.
JT was PI and conceived the original BRISSKit concept and design building on early use cases developed with NH & colleagues in the NIHR Cardiovascular Biomedical Research Unit. JT managed the project through subsequent stages at the University of Leicester including leading on all strategic, funding, partner, contractor and institutional liaison in particular with Jisc, University Hospitals Leicester NHS Trust and the UK National Institute for Health Research.
MN & NH were heavily involved in the technical design of BRISSKit. PB was centrally involved in strategic development of the BRISSKit project.
OB, JT, RW, SI & PB prepared and reviewed the manuscript.
The bulk of the development work took place at the University of Leicester (PI J.Tedds) and was supported by Jisc under the HEFCE University Modernisation Fund - Shared services and the cloud programme 2011:2012 with additional funding during 2012:2013 and 2014:2015 from Jisc and the University of Leicester. OB is funded under a strategic award from MRC and Wellcome Trust for the ALSPAC project [102215/Z/13/Z]. The University of Bristol component of the work described in this article is funded as a central element of the research program of the Data to Knowledge (D2K) Research Group, jointly supported by funding from: the European Union’s Seventh Framework Programme BioSHaREEU [261433] (Biobank Standardization and Harmonization for Research Excellence in the European Union) and BBMRI-LPC [313010] (Biobanking and Biomolecular Resources Research Infrastructure - Large Prospective Cohorts). MRC: the Welsh and Scottish Farr Institutes, MRC funded E-Health Informatics Research Centres (EHIRCs) [MR/K006525/1; MR/K007017/1]. Wellcome Trust & MRC: 58FORWARDS [108439/Z/15/Z] (The 1958 Birth Cohort: Fostering new Opportunities for Research via Wider Access to Reliable Data and Samples).
The authors acknowledge the contributions of a wide range of academics and support staff at the University of Leicester during this work including in the College of Medicine, Biological Sciences and Psychology, IT Services, Research and Enterprise Division and Library. We also thank the many NHS staff at University Hospitals Leicester NHS Trust including Chris Greengrass, Tim Skelton, John Clarke, Andy Carruthers, David Rose, Richard Bramley, Alison Goodall, Nilesh Samani, David Wynford-Thomas, Kevin Schurer, Anthony Brookes, Mary Visser, Robert Feakes and a range of other partners, collaborators and external contractors involved; early pilot evaluators led by Dr Tito Castillo at Great Ormond Street Hospital, University College London, and at the School of Cancer Studies, University of Birmingham, led by Paul Mason. We highlight the long standing and much valued insight and support from Peter Knight, Deputy Director at the Department of Health; John Milner, Simon Hodson, Rachel Bruce, Daniela Duca, Catherine Grout, Martin Hamilton and colleagues at Jisc; and Neil Beagrie and Daphne Charles at Charles Beagrie Ltd.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 02 Aug 16 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)