Keywords
developmental gene regulatory networks, sea urchin, dgrn, patterning, morphogenetic cassetes,
developmental gene regulatory networks, sea urchin, dgrn, patterning, morphogenetic cassetes,
Developmental gene regulatory networks (dGRNs) describe the sequential regulatory changes that specify and diversify the cells of an embryo. The genes included in dGRNs encode transcription factors, components of signal transduction pathways, and often effector genes as markers of differentiated cell states. Models of dGRNs are assembled on the basis of experimental perturbations of an embryo’s developmental program and are valuable for explaining how spatial and temporal information is encoded in a multicellular organism’s genome. dGRNs have the potential of providing a causal understanding of how upstream specification controls downstream events (i.e. differentiation or cell biological functions). As such, a dGRN can serve as a tool for developmental and cell biologists alike. This article describes how dGRNs are assembled using the sea urchin embryo as a model and includes recent insights into sea urchin development that have benefitted from the assembly exercise.
The early specification events of the sea urchin embryo have been extensively documented, resulting in increasingly well-understood dGRNs for each cell type. Sea urchin development is relatively simple, easy to observe, and experimentally tractable, and experimental outcomes are rapidly obtained. Prior to genome sequencing, a provisional sea urchin endomesodermal dGRN was assembled starting with a small number of transcription factors and signaling inputs1. In 2006, the sea urchin genome was sequenced2, and since that time, labs across the globe have added to and reinforced an understanding of the regulatory linkages in the dGRNs through gastrulation3–9. In contrast to typical “hairball” or “interactome” networks, where hypothetical interactions are based on statistical inference, each linkage of the sea urchin dGRN is based on several experimental approaches, spatial and temporal validation, and in many cases cis-regulatory analyses to confirm a direct regulatory connection. This effort has provided many valuable insights into network function and has led to novel explorations into mechanisms of sea urchin development. We begin by summarizing experiments that demonstrated how the sea urchin dGRN was, and continues to be, assembled. Then we briefly review how dGRNs have been used as tools to understand how embryonic patterning works, how morphogenesis is controlled, and how evolutionary processes have modified dGRNs.
GRNs reflect the relationship between genes in a system. In a graphic depiction of GRNs, the expression of a transcription factor or a component of a signal transduction pathway is represented as a node. The edges or connections between nodes reflect the regulatory relationship between nodes over time. In GRN models, an upstream regulator is drawn with an output that either activates or represses expression of a downstream gene. At any given time after development begins, many transcription factors are expressed and are under regulatory control by transcription factors upstream of them. Over time, this produces a regulatory network with many properties. At any given time, a cell in an embryo is controlled by the unique regulatory “state” of its GRN. All identical cells will tend to be regulated by the same GRN state, and as cells diverge from one another, their regulatory states change. Signaling molecules produced by one cell pass to adjacent or nearby cells where their inputs alter GRN states in recipient cells. Within a GRN state, it is possible to tease apart relationships of transcription factors and identify sub-circuits designed to accomplish the tasks of development. Among the tasks uncovered in the sea urchin dGRN are a number of properties that are commonly seen when network systems are studied in detail10–12. For example, a “double-negative gate” (repression of a repressor) was identified to initiate specification of a specific cell type4,13. “Spatial exclusion” sub-circuits were found in cells at the time these cells diverged from one another toward differentiation into distinct cell types; in each of the two nascent cell types, there was a sub-circuit to exclude the other14. “Community effect” signaling was identified such that Nodal reinforced Nodal signaling in neighboring ectodermal cells to maintain Nodal signal production in that community of ectodermal cells11,15. “Feed-back sub-circuits”, in which a downstream transcription factor feeds back to maintain activation of an upstream transcription factor, and “feed-forward sub-circuits”, in which an upstream transcription factor feeds forward to activate multiple sequential downstream steps, were found. Feed-back and feed-forward sub-circuits tended to stabilize regulatory states4 and contribute to the unidirectional trajectory of development. Sub-circuits just upstream of differentiation were found that drive the system forward and contribute to the activation of genes necessary for differentiation and cell biological function (e.g., 6,16–18). These and other sub-circuits provide dGRNs with modular design features that control explicit functions. As details of dGRN topology were identified, they were shown to contribute impressively to a growing understanding of developmental mechanisms in many systems and are central to research in sea urchin development as a consequence.
When an early draft of the Strongylocentrotus purpuratus genome became available, it provided an opportunity to identify hundreds of transcription factor genes and molecules of signal transduction pathways (Figure 1). Each candidate development regulatory gene (transcription factors and signal transduction pathway gene) was assayed by quantitative polymerase chain reaction (qPCR) and whole-mount in situ hybridization (WMISH) to establish spatial and temporal expression patterns throughout early development19–25. Of the large number of transcription factors identified, those that were spatially or temporally distinct (or both) in expression in the early embryo were chosen for detailed efforts to define the distinct regulatory states of the embryo at a number of time points, beginning with fertilization and continuing through gastrulation. Ubiquitously expressed genes initially were excluded from the study with the notion that they were less likely to be involved in developmental regulatory decisions. Perturbation analyses of each identified signaling molecule and transcription factor then established linkages between nodes. Morpholino antisense oligonucleotides knocked down one transcription factor or signal, and analyses by qPCR, WMISH, or nanostring (or a combination of these) assayed the effect on other genes expressed at the same time, or in the same cell type, or both. This established a hierarchical relationship between the transcription factors, signals, and the genes encoding them in each cell type over time. The regulatory interactions were assembled into a network model by using BioTapestry as a platform for visualizing network topologies26,27. Many of the interactions were validated by using cis-regulatory analysis to determine direct binding of the transcription factor to specific sequences in the regulatory DNA of downstream genes. With this logical approach, hundreds of experiments were performed to establish sea urchin dGRN states in each emerging cell type up to gastrulation, and current efforts continue to extend the analysis to later time periods in development (e.g., 18,28–30).
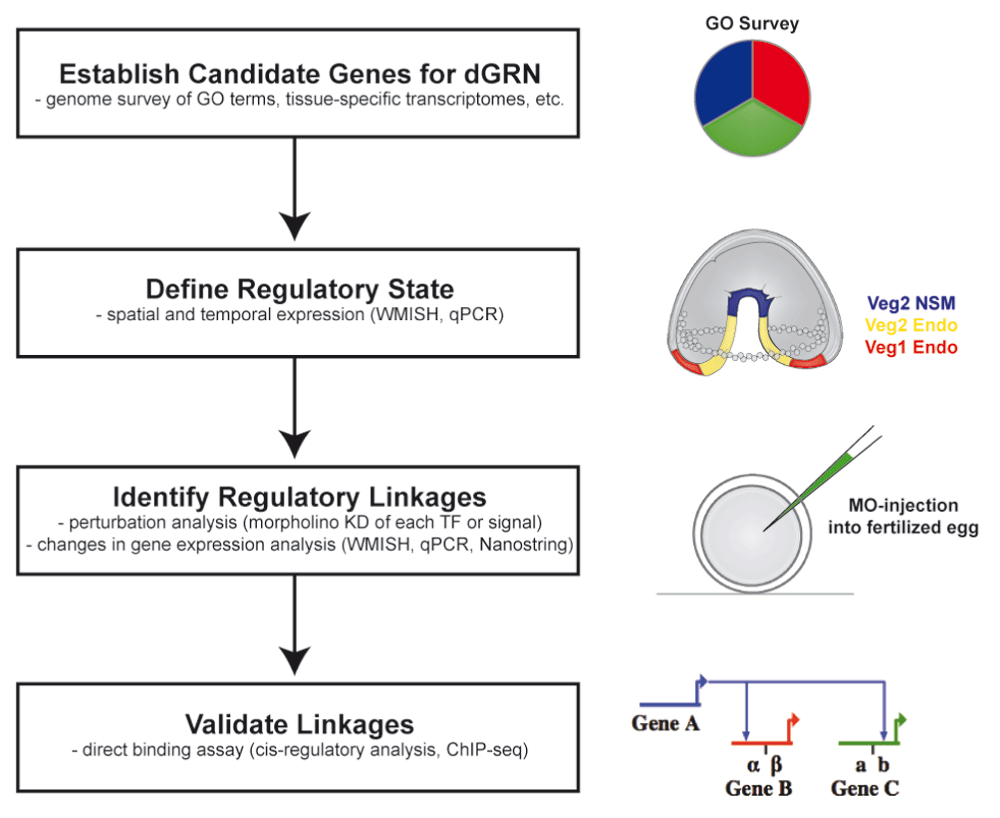
The process begins with identification of candidate molecules. Candidates for the sea urchin dGRN were defined as transcription factors or signal transduction pathway members that were expressed in spatiotemporally specific patterns in the embryo. The regulatory linkages were established by conducting perturbation analyses in which one candidate was perturbed and asking how its loss affected expression of other candidates. These established a preliminary dGRN model. That model was challenged in many ways, including testing predictions that gene A activated gene B and gene C through cis-regulatory analysis.
The term “patterning” describes processes that establish the body plan of an organism. Patterning processes provide “positional information” enabling cells to know their location in an embryo. These processes direct morphogenesis, and they provide short-range and long-range signaling inputs to accomplish construction of a three-dimensional plant or animal (Figure 2). Each of these processes is driven by dGRNs. The sea urchin embryo begins patterning almost immediately after fertilization. By the 16-cell stage, zygotic transcription plus maternal inputs defines at least three distinct dGRN states (Figure 2). At this time, the future mesoderm and endoderm are combined as an endomesoderm network state. A Delta-Notch signal separates progeny of these cells into mesoderm (cells that receive the Delta signal) and endoderm (endomesoderm progeny that do not receive the Delta signal)31–33, and further specification plus signaling subdivides the initial endomesoderm GRN state and ectoderm GRN state until there are at least 15 different cell types recognizable by early gastrulation (Figure 2)7,34.
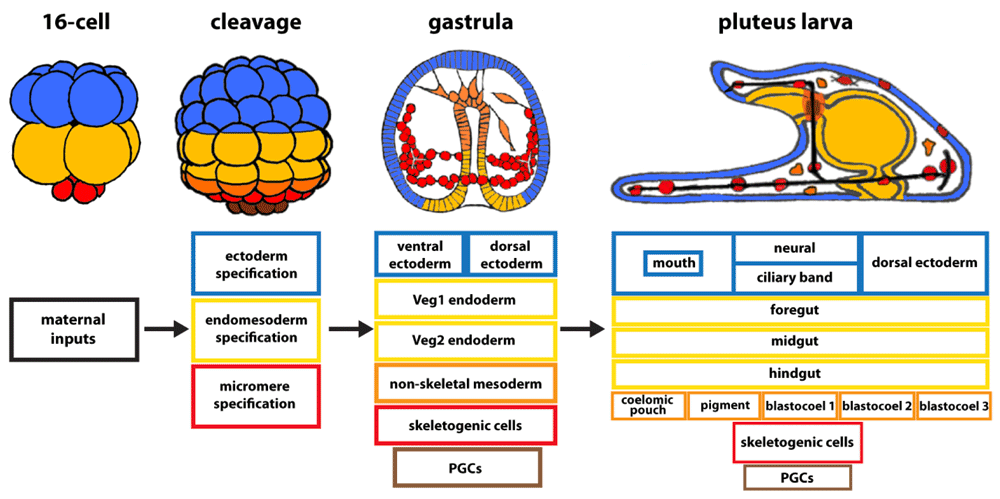
Over the 24 hours from fertilization to the pluteus larval stage of Lytechinus variagatus, the number of developmental gene regulatory network (dGRN) states increases until there are more than 15 cell types in the early larva.
Signaling is integral for the patterning mechanisms that organize multicellular cell and tissue types. As specification progresses, signaling establishes the three axes of the embryo. Early patterning signals divide the embryo into regions along the animal-vegetal axis, established by asymmetrically localized maternal information, followed by a series of Wnt signaling events that progressively diversify cell identities along the anterior-posterior axis35–41. The anterior-posterior axis of the larva is approximately identical to the animal-vegetal axis of the egg. At about mid-cleavage, the dorsal-ventral (D-V) axis is specified through Nodal and bone morphogenetic protein (BMP) signaling15,42,43, and toward the end of gastrulation, the right-left axis is established, again using Nodal and BMP and adding Hedgehog as a contributing signal16,44–47. Thus, by the end of gastrulation, signaling inputs plus a progression of transcription factor activations establish dGRN states for each cell type in the embryo, and even within the same germ layer, patterning inputs provide localized cell identities.
Patterning continues to play a key role in organizing structures in the larva. As an example, a biomineralized skeleton provides the shape to the pluteus larva. To pattern that skeleton, signals from the ectoderm are received by the mesodermal skeletogenic cells, enabling them to position themselves correctly and to synthesize the calcium carbonate biomineral in the correct pattern48–51. Ectodermal signals are supplied from specific locations requiring the ectoderm to be patterned in advance. Ectodermal positional information leads to specification of two lateral patches of ectodermal cells specialized in secretion of vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF)5,6,8,49,50. To provide that positional information, orthogonal bands of ectoderm are specified by even earlier signals. One of the ectodermal bands, the ciliary band, is specified at the boundary between the dorsal and ventral ectoderm as a consequence of Nodal (ventral) and BMP (dorsal) signaling15,42,52,53. BMP is synthesized in the ventral ectoderm and is transported, probably using Chordin as a chaperone, to induce dorsal ectoderm specification29,53,54. The ciliary band then forms between the dorsal and ventral ectodermal territories and is further subdivided29. The other ectodermal band, referred to as the border ectoderm, is specified in posterior ectoderm, just above the endoderm and orthogonal to the ciliary band separating the D-V ectoderm compartments. Wnt5 signaling from the endoderm is reported to induce the border ectoderm band in Lytechinus55,56, although a different Wnt is thought to be responsible in S. purpuratus56, and that band also receives input from the Nodal-BMP signaling as seen in both Lytechinus and Paracentrotus30,55. The site where the ciliary band and the border ectoderm band intersect becomes the signaling center that produces VEGF and FGF, both of which are necessary for initiation and growth of the skeleton49–51. Other signals also provide patterning inputs into skeletogenesis58. These data demonstrate that, in an organism, cells can be specified independently for long periods of time to seemingly establish independent dGRNs but that, at a later time, through signaling their dGRNs functionally intertwine once again. A good example is skeletal patterning, where signals from specific locations in the ectoderm provide patterning information and growth factors that direct the behavior of the mesoderm.
Morphogenetic control circuitry is another area where the dGRN is valuable (Figure 3). Each morphogenetic change incorporates multiple cell biological functions: changes in adhesion, motility, directionality, polarity, and so on. Perturbations that assess which transcription factors control individual cell’s biological properties have been valuable in dissecting details of those processes17,18,51,56,59–62. In an epithelial-mesenchymal transition (EMT), for example, five different dGRN sub-circuits control de-adhesion, motility, cell polarity, cell shape change, and invasion components of the EMT process61.
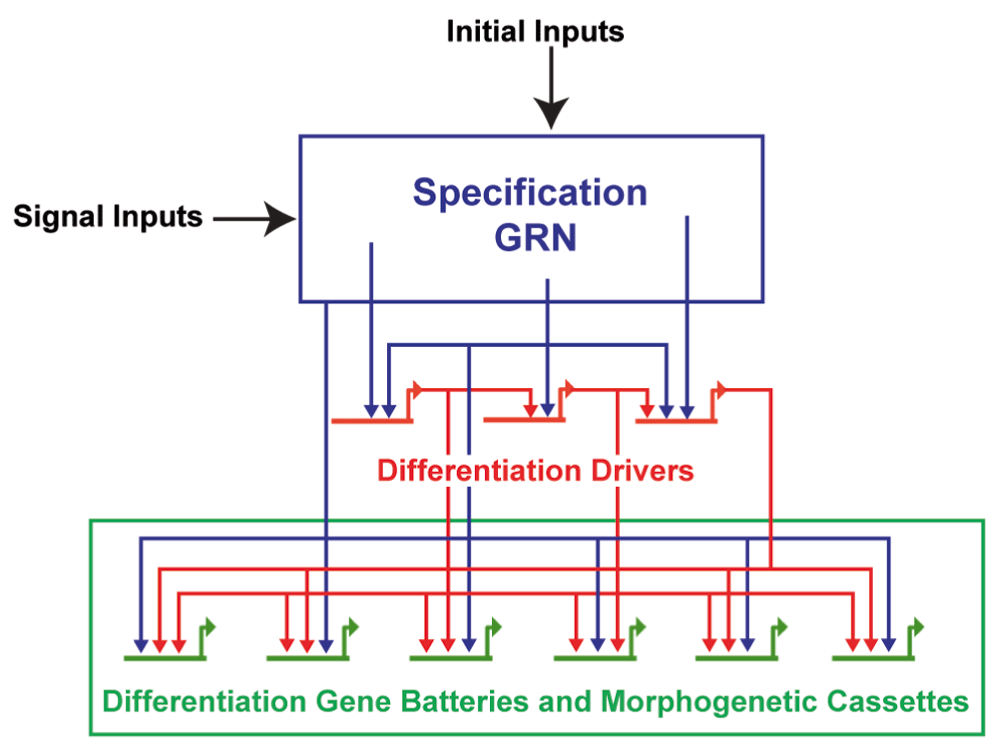
Maternal inputs initiate specification. As cells divide, signaling becomes increasingly used to shape the specification sequence of each cell type. Toward the end of that process and proximal to differentiation, transcription factor sub-circuits drive expression of genes involved in differentiation and in morphogenesis.
Later in development, a specific feed-forward sub-circuit controls the directed migration of the presumptive primordial germ cells18. Identification of such sub-circuits can then be used to penetrate the cell biology of each component process for each of the cell types (or organs) that have been defined in the larva. Along these lines, the circuits controlling ciliary band specification were identified, allowing for further analysis of the substrate for neural differentiation29. Yet, other circuits that control myogenesis of the larva were recently identified28. In each of these cases, the dGRN was valuable in gaining access to differentiation genes and effector genes ultimately controlled by that developmental GRN (Figure 3).
A third valuable reason for studying dGRNs is to learn how evolutionary processes have changed dGRNs during species diversification. The sea urchin dGRNs are modeled primarily from data generated from three species of euechinoid sea urchins that are separated from one another by about 50 million years or less63. Examination of more distantly related species reveals how rewiring of dGRNs correlates with evolutionary changes in morphology and cell behavior. For example, an examination of a cidaroid sea urchin, separated from the euechinoids by more than 255 million years, revealed significant rewiring of the dGRN that specifies the larval skeletogenic lineage64,65. In euechinoids such as S. purpuratus or Lytechinus variagatus, the skeletogenic lineage is specified early during cleavage stages, and after a species-specific number of cell divisions, all of the skeletogenic cells undergo EMT before gastrulation of the other mesodermal and endodermal lineages. In the cidaroid, Eucidaris, the number of skeletogenic cells varies from embryo to embryo, and the EMT of these skeletogenic cells does not occur until much later in development relative to Lytechinus66. In Strongylocentrotus and Lytechinus, the skeletogenic lineage is specified by a core set of genes, including Alx1, Tbr, and Ets1, which are transcriptionally activated specifically in this lineage by unlocking a double-negative derepression sub-circuit controlled by Pmar1 and HesC4. In the cidaroid, Eucidaris, this double-negative derepression sub-circuit appears to be completely missing, and the spatiotemporal expression patterns of Alx1, Tbr, and Ets1 are different as well64. Thus, extensive rewiring of the dGRN occurs in sketogenic cell lineage, and morphogenesis of the skeletogenic cells differs, yet both modern sea urchins and the pencil urchins produce similar larval skeletons.
Comparative dGRN analysis can also reveal what aspects of specification are highly conserved during evolution. For example, experiments perturbing the function of Alx1 in euechinoids, cideroids, and sea cucumber have shown that Alx1 has a conserved role in promoting larval skeletogenesis64,67. At even larger evolutionary distance within echinoderms, valuable insights have been obtained through comparisons of skeletal, gut, and neural specification in sea stars compared with sea urchins. In each case, aspects of central dGRN circuitry were very similar despite about 500 million years of separation from the common ancestor68–70. dGRNs can also be used as a tool for understanding how circuitry has been co-opted during evolution. In a recent study, for example, it was learned that a conserved feed-forward sub-circuit involving Pax6, Six3, Six1/2, Eya, and Dach1 controls expression of the signal necessary for homing of primordial germ cells to the future gonad18. That circuit is very similar to the feed-forward circuit that controls specification of the Drosophila eye and vertebrate muscle, suggesting that such circuits may in some cases evolve as units of function, in this case by providing a feed-forward device for directed cell migration.
Sea urchin dGRNs describe the sequence of specification of all cells in the embryo up to the end of gastrulation. dGRN topology models produced in BioTapestry (http://sugp.caltech.edu/endomes/) record the current status of the network in S. purpuratus. Much more than a graphic description, it reflects a large number of experiments, where each connection is supported by multiple tests of the hypothesis that the expression of gene A activates or represses gene B. In its current form, the sea urchin dGRN includes more than 100 transcription factors and a number of signaling pathways, and in most cases multiple laboratories have validated each connection in S. purpuratus, and most are the same in Lytechinus and Paracentrotus.
With a high level of confidence in the structure of the sea urchin dGRNs, they became useful as a tool for exploring many other developmental questions. Here, we describe how the dGRNs have been used to inform patterning mechanisms, especially those necessary to produce the larval skeleton. We show how they have been useful in gaining a greater understanding of an EMT and a directed cell movement mechanism, both components of morphogenesis. We also describe how dGRNs are used as tools for discovery of evolutionary relationships.
The growth in our understanding of dGRNs has provided ways to address many unanswered questions. The ability to trace the entire specification trajectory of a cell type until it terminally differentiates is now a realistic goal. That ability has enormous power because it allows one to interrogate, dissect, and understand how that cell type arises and how it works in detail. This will be valuable in understanding the entire history of neurogenesis, for example, and in determining the mechanisms by which neurons diversify toward different neurotransmitter cell types. Other cell types can be followed with the same goal. dGRNs will also help us to understand how the upstream circuitry controls other morphogenetic movements in the early embryo. Thus, the information in the dGRN is useful both for gaining an intrinsic understanding of how developmental control circuitry works and as a tool for understanding patterning, morphogenesis, and evolutionary change.
Support for this article was provided by grants from the National Institutes of Health (RO1-HD14483, PO1-HD03705 to DRM).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 22 Feb 16 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)