Keywords
Antibiotic resistant bacteria, Xenorhabdus bacteria, novel antimicrobials
Antibiotic resistant bacteria, Xenorhabdus bacteria, novel antimicrobials
In this version, a few typographical errors have been corrected. The contributions of Daniel Masiga, previously only recognised in the Acknowledgments section of version 1, significantly increased. Thus, he is now recognized as a co-author of the research paper. Also, the new Acknowledgments section has been updated to conclusively reflect the role various parties played in bringing this research, over its 8-year journey, to light.
To read any peer review reports and author responses for this article, follow the "read" links in the Open Peer Review table.
Antibiotic resistant bacteria, otherwise known as “superbugs”, are an imminent threat to every existing healthcare system as they could obviate current clinical antibiotics and thereby retrogress humanity to that dark age of lethal sore throats1. Of note is methicillin-resistant Staphylococcus aureus (MRSA). In this study, we examine the antimicrobial activity of Xenorhabdus griffiniae fermentation media against MRSA.
MRSA not only causes human diseases such as mastitis, chronic open wound infections and endocarditis, but also livestock diseases such as mastitis in dairy cattle and lameness in poultry and rabbits2, that together result in economic losses of billions of dollars to the agricultural sector3. In both humans and animals, an MRSA infection can quickly turn lethal. This is because MRSA is resistant to two antibiotic classes, beta lactams and macrolides, and only lipopeptides and glycopetides remain effective4; this development has contributed in large part to the rise of the vancomycin-resistant S. aureus superbugs. A recommended solution1 to the superbug conundrum is to develop novel classes of antibiotics that can replace those to which disease-causing bacteria have mutated to resist their inhibitory effect.
A potential source of novel anti-MRSA antibiotics are Xenorhabdus bacteria5 that naturally dwell in the guts of Steinernema roundworms. These 1 mm-long6 roundworms are found in soils worldwide7 and live by infecting and killing insects such as moths, caterpillars and weevils. The Xenorhabdus bacteria carried in their gut aid this insect killing lifestyle. Explicitly, Steinernema roundworms enter an insect body and release Xenorhabdus bacteria that secrete insecticidal toxins that quickly kill the insect8. To secure this rich food source for the roundworms only, the Xenorhabdus produce an armory of antibiotic compounds that effectively destroy competing soil fungi and microorganisms9, a mechanism that has been demonstrated as having medical potential against human diseases8,10–14. Each Xenorhabdus species has been demonstrated to produce its own unique array of antibiotics11 and this is prompting new studies on the classes of antimicrobial compounds produced. For example, X. cabalinasii JM26 from Jamaica led to the discovery of nemaucin, a novel and highly potent antibiotic compound against methicillin-resistant S. aureus15.
Politically, “superbugs” are today’s top global health issue exemplified by the 2016 United Nations High Level General Meeting’s agenda being antibiotic resistant bacteria: this is only the fourth time in its 80-year history that a health issue has been the reason for this annual meeting. Technically, superbugs have been detected in all 114 countries recently surveyed with the number of pan drug resistant superbugs, those immune to every antibiotic available, on the rise1. Accordingly, Kenyan prevalence levels of MRSA have been steadily increasing16,17.
Previously18 we demonstrated that Xenorhabdus bacteria from Kenya can produce antibiotics against MRSA. However, the identity of the species, the antibiotic classes it produces and their inhibitory concentrations remain unknown. Here, we elucidate the specific molecular identity of Kenyan Xenorhabdus isolates and determine the efficacy of produced antimicrobial compounds against MRSA. Our findings highlight a novel antibiotic class designated “mursamacin”19 obtained from Xenorhabdus bacteria found in Kenyan soils, which is highly active against methicillin-resistant S. aureus.
MRSA strain 133 cultures were obtained as a gift from Dr. John Ndemi of the Kenya Medical Research Institute, Centre for Microbiology Research, Nairobi, Kenya. Pure nematode cultures of Steinernema roundworm isolates were obtained from the nematode culture collection of Horticulture Research Institute Thika, Kenya.
Using the indirect haemolymph method20 X. griffiniae strains XN45 and L67 were isolated from Steinernema sp. Scarpo and Steinernema sp. L67 nematodes respectively. The isolates were cultured on Xenorhabdus differential media NBTA, composed of nutrient agar (Himedia) supplemented with 0.0025% (w/v) bromothymol blue (Sigma-Aldrich) and 0.004% (w/v) 2,3,5 triphenyl tetrazolium chloride (Sigma-Aldrich)20. Identification of the bacteria as Xenorhabdus was based on the presence of the following characteristics: swarming motility on NBTA of 1% (w/v) agar concentration; swimming motility on NBTA of 0.5% (w/v) agar concentration; and yellow green colony pigmentation on NBTA5. Specific identification of the bacteria was done by multi locus sequence typing of the 16s rRNA, serC and recA genes (See Dataset 1).
Total DNA extraction from the bacterial strains was done using FastDNA®SPIN Kit for Soil (MP Biomedicals, USA). Isolation of a 1397 base pair(bp) 16s rRNA gene fragment was done by Polymerase Chain Reaction (PCR) with primer sequences (Inqaba Biotech) as follows: (27f-AGA GTT TGA TCA TGG CTC AG) and (1391r-ACG GGC GGT GTG TGC)21. The genes were amplified in a 25 μl reaction volume containing final concentrations of 0.5 U Q5 DNA polymerase (New England Biolabs, USA), 200μM each dNTP, 2mM MgCl2, and 0.05μM of each primer. Cycling conditions were set at 98°C for 30 s, 40 cycles of 98°C for 30 s, 42°C for 15 s (first 20 cycles) and 47°C for 15 s (final 20 cycles), 72°C for 1 min and a final extension of 72°C for 2 min (MJ Research PTC-100, USA). Isolations of 670bp serC and 400bp recA gene fragments were done by PCR with primers (recA-FW CCA ATG GGC CGT ATT GTT GA) and (recAREV-TCA TAC GGA TCT GGT TGA TGA A) and (serCF-CCA CCA GCA ACT TTG TCC TTT C) and (serCR- AAA GAA GCA GAA AAA TAT TGC AC) respectively22. They were amplified in a 50 μl reaction volume containing final concentrations of 2 U MyTaq® DNA polymerase (Bioline, USA), 200μM of each dNTP, 3mM MgCl2, and 0.4μM of each primer. For both, cycling conditions were set at 95°C for 1 min, then 40 cycles of 95°C for 15 s, 52°C for 15 s, 72°C for 40 s, with a final extension of 72°C for 5 min (Thermo Scientific Arktik, USA).
PCR products were visualized on 1.2% (w/v) agarose gels stained with ethidium bromide at final concentrations of 0.5 μg/ml. Typical electrophoresis conditions were 4V/cm for 72 min. Expected bands were excised and purified with Quick Clean II Gel extraction kits® (Genscript, USA). Products were outsourced for sequencing (Macrogen, Netherlands), and obtained sequences were quality checked, assembled and poor quality base calls trimmed in BioEdit23 and MEGA624 software suites.
Phylogenetic reconstruction was performed using a multi locus concatenate of 16s rRNA, serC and recA genes sequences, that jointly constituted 2076 positions. A dataset of n=13 (1= from this study and 12= public databases) was used that contained the 11 strains of Xenorhabdus and a Photorhabdus luminiscens (the out-group sequence) that had public database sequences of all three genes (see data files). Database sequences were checked for quality and ambiguous nucleotides resolved in the MEGA6 software suite24. Multiple sequence alignments were performed in the same suite using the MUSCLE algorithm25. The evolutionary history was inferred by the ML method based on the Generated Time Reversible (GTR) Model (500 bootstraps). Initial trees for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The analysis involved 13 nucleotide sequences. All positions containing gaps and missing data were eliminated resulting in a total of 2076 positions used in the reconstruction. Xenorhabdus DNA sequences generated in this study were deposited in the DNA databank of Japan with the partial 16s rRNA gene sequences of X. griffiniae L671, L672, L673, L675, XN45 assigned the following accession numbers respectively; AB987698.1, AB987700.1, AB987701.1, AB987699.1, AB987697.1. The partial recA, serC gene sequences of X. griffiniae L67 and XN45 were assigned LC096094, LC096092 and LC096093, LC096091 respectively. Percentage sequence similarities between gene sequences from this study and other 16s rRNA, recA and serC gene database sequences was determined using blast searches26.
Fermentation was done using X. griffiniae XN45 bacterial cultures. Multiple colonies (2–3) of an individual isolate were selected, inoculated into 5 ml of Luria Bertani (LB) media containing 1% tryptone, 0.5% yeast extract and 1% NaCl (w/v) and incubated on a rotatory shaker at 150 rpm at 33°C for 24 h. These served as 1% (v/v) starter inocula. Sterile LB media (500 ml) was dispensed into sterile 1-L Erlenmeyer flasks, with starter cultures (5 ml) thereafter inoculated and LB media incubated at 150 rpm at 33°C for 355 h, 180 h and 108.5 h. LB with no inoculum was also incubated to serve as a control for sterility. After fermentation, cells were removed by centrifugation of broths at 20,000 g for 25 min at 4°C (Beckman Avanti J-25, USA) followed by decanting cell free supernatants (cfs). These were heat-treated by autoclaving at 121°C and 15 p.s.i for 20 min to yield a sterile heat stable fraction of the whole broth extract (antibiotic) that was designated “mursamacin”. These were stored at 4°C until use.
The broth macro dilution assay was prepared as previously described11 with modifications, using antibiotics obtained from each of the fermentation durations (180.5 h and 355 h) as the antibiotic and MRSA as the test bacterium. MRSA overnight cultures were inoculated into each dilution, at a final concentration of 2.3×104 cfu/ml, and then incubated for 18 h at 37°C without agitation. The following controls were included in every replicate: negative control of 2× LB media inoculated with bacteria without antibiotic; sterility control of 2× LB media with no inoculated bacteria; and sterility control of undiluted antibiotic with no inoculated bacteria.
After incubation, turbidity of each dilution was measured by determining the A600nm (See dataset files). This was used in the following formula, modified from Houard, Aumelas13 that included a correction factor for inhibition by the broth media to calculate the percentage growth inhibition of bacterial cultures by an antimicrobial.
Where g = A600nm of bacteria in broth culture without antibiotic, and gx = A600nm of bacteria in broth culture with antibiotic.
To extract an organic heat stable mursamacin class of antibiotics, cfs from a 108.5 h fermentation reaction were lyophilized to yield a yellow powder, whose measured amounts were dissolved in known volumes of methanol, and vortexed for 2 min to yield a dark orange methanol extract. These were centrifuged at 20,000 g for 13 min at room temperature to separate liquid methanol extracts from insoluble residues. Methanol extracts were pipetted into sterile 1.5 ml tubes while pelleted solids were aseptically air dried and then incubated at 37°C overnight to evaporate residual methanol. To provide a negative control, the same procedure was repeated for lyophilized powders of the fermentation media. The difference in weight of powder before and after methanol extraction was then measured to determine the concentration of total dissolved compounds in the methanol extracts.
This was modified from a previously described method27. To determine the minimum inhibitory concentrations (MICs) against MRSA, 100 μl of known concentrations of organic heat-stable fractions of mursamacin antibiotics were dispensed into sterile 96-well micro-titre plate starting wells. These were left in a biological safety cabinet to evaporate methanol resulting in visible orange solid residues; each was then re-dissolved in 200 μl RPMI media supplemented with 5% (v/v) LB28. Every other well along each row was filled with 100 μl RPMI media supplemented with 5% (v/v) LB; 100 μl of starting well mixture was then dispensed to the subsequent well resulting halving the antibiotic concentration. This was repeated for all wells along the rows resulting in a 2-fold dilution series. MRSA inocula (10 μl) were used that had been previously prepared from plate cultures dissolved in physiological saline to a turbidity of 0.5 Mcfarland standard (ca. concentration= 2.6×106 cfu/ml), then subjected to 10-3 dilution in RPMI media supplemented with 5% (v/v) LB. A positive control row of Daptomycin and negative control of methanol extract of LB media only was incorporated in every replicate. Plates were incubated at 37°C for 21 h without agitation. Experiment was performed in 10 replicates in three reproductions.
To identify the compounds contained in the organic heat-stable fraction of mursamacin antibiotics, analytical reverse phase chromatography of the methanol extract was performed as previously described13 and modified to use of a c18 column (Aglient Zorbax Eclipse Plus C18; 3.5um, 4.6 ×100 nm) under isocratic conditions of 60% acetonitrile and uv detection of 224 nm.
The Xenorhabdus isolates identified were most closely related to Xenorhabdus griffiniae (Figure 1). The apt thresholds for Xenorhabdus species identification based on sequence similarities are currently considered to be 98.65% and 97% for 16s-rRNA and for both recA and serC gene fragments respectively29,30. In this study, sequences similarities to X. griffiniae strains, including type strains, were 99.52%, 98.57% and 97.68% for 16s rRNA, recA and SerC respectively (See data files)

The tree was based on a concatenate of ss-rRNA, recA, and serC gene sequences and was reconstructed with a general time reversal model and test of phylogeny of 500 bootstrap replicates. X. griffiniae isolated from this study (red triangle) clustered in the X. griffiniae clade (red), and was most closely related to X. griffiniae from Malaysia.
Xenorhabdus griffiniae has only previously been isolated from Indonesia31, Malaysia32 and South Africa33; therefore our data strongly suggest a new strain of X. griffiniae originating from Central Kenya.
The growth of methicillin-resistant S. aureus was inhibited when cultured in X. griffiniae cell-free supernatants (cfs) that we termed ‘mursamacin’ (Figure 2). Furthermore, cfs were obtained by various X. griffiniae fermentation durations and had the following percentage of growth inhibition against methicillin-resistant S. aureus at neat concentrations: 81% (cfs from 180 h ferment), 94% (cfs from 355 h ferment) (Figure 3). In both instances, cfs were heat-sterilized by autoclaving and pH adjusted to that of the control (6.6–7.0), indicating that inhibition was due to heat stable compounds contained therein.
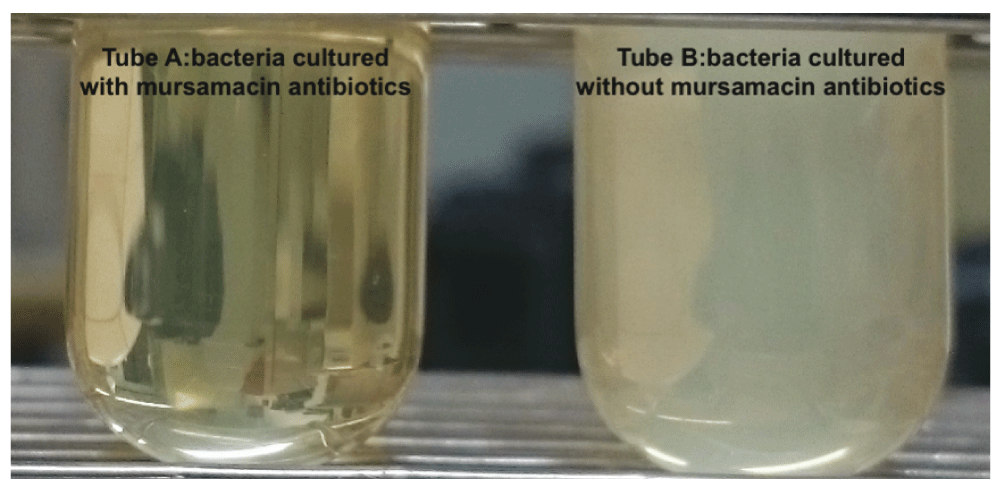
MRSA cultures were incubated with (tube a) and without (tube b) mursamacin antibiotics respectively. A clear tube denotes no bacterial growth while turbid tube denotes bacterial growth.

The longer fermentation duration (355 h) produced antibiotics that were generally more inhibitory to methicillin-resistant S. aureus. Dotted line graphs represent linear equations derived from the raw inhibition values. The high R-squared values demonstrate that the concentration of the antibiotic was predominantly responsible (97–98%) for the level of percentage growth inhibition.
Previous studies9,11–13,34,35 have demonstrated that Xenorhabdus bacteria are prolific antimicrobial producers with each species producing its unique array of antibiotics that often contained novel compounds. Consistent with these reports, our data demonstrate unprecedented evidence of heat-stable X. griffiniae antimicrobials and further suggests that their production is affected by how long X. griffiniae is cultured in fermentation media.
An organic heat-stable fraction of mursamacin antibiotics inhibited methicillin-resistant Staphylococcus aureus at a standardized concentration of 8.25 μg/ml while its negative control- organic extract of autoclaved fermentation media only- displayed growth at all concentrations confirming this inhibition as due to antibiotic compounds only (Table 1). However, this concentration was 17-fold higher than the positive control Daptomycin that gave 0.5 μg/ml.
Currently, there are two major clinical antibiotics with inhibitory concentrations against methicillin-resistant S. aureus that are considered effective: Daptomycin (0.5 μg/ml) and Vancomycin (2 μg/ml)4. On the other hand, the larger majority today’s clinical drugs have inhibitory concentrations against methicillin-resistant S. aureus considered that are considered ineffective; Azithromycin (128μg/ml), Amoxicillin/Clavulanic acid (64 μg/ml), Cefriaxone (64 μg/ml) and Erythromycin (32 μg/ml) and Imipenem (16 μg/ml)4. Yet our data demonstrate a heat stable class of compounds with an inhibitory concentration of 8.25 μg/ml. This strongly suggests that antimicrobials contained in this class are even more potent against methicillin-resistant S. aureus, as pure compounds.
Further high performance liquid chromatographic analysis revealed that this fraction contained two major compounds, eluted at 1.862 min and 2.775 min and absorbed at 224nm when dissolved in methanol (Figure 4). Previous studies have characterized classes of organic Xenorhabdus antibiotics that were highly effective against S. aureus11,36 and had peaks absorption ranges from 200–230nm15. Of note, the PAX lipopeptides10 isolated X. cabanillasii and X. nematophila were highly effective against MRSA at concentrations of 0.5 μg/ml. Yet in contrast to our results, no class has been characterized as heat stable and isolated from X. griffiniae.

Top chromatograph represents the solvent only while bottom chromatograph is of solvent containing organic mursamacin antibiotics. Two dominant compounds, indicated by the red arrows, were detected in this fraction.
In conclusion, we demonstrate that X. griffiniae antibiotic compounds termed “mursamacin” contained an organic heat stable fraction are highly effective against methicillin-resistant S. aureus and antimicrobial activity seems to be attributed to two dominant uncharacterized compounds. This may offer a founding stone for further development of clinical drugs from X. griffiniae, giving hope to thousands of patients affected by methicillin-resistant S. aureus infections.
F1000Research: Dataset 1. Raw data of antibiotics from soil dwelling roundworms of Central Kenya inhibiting methicillin resistant Staphylococcus aureus, 10.5256/f1000research.9652.d13696637
Accession numbers of sequences generated from this study are: AB987698.1, AB987700.1, AB987701.1, AB987699.1, AB987697.1. ,LC096094, LC096092 and LC096093, LC096091.
RMA, PNN, LNN conceived the study and carried out the research. NA designed the experiments and supervised the study. DM supervised the study and provided a laboratory, equipment and reagents. RMA wrote the manuscript. All authors were involved in the revision of the manuscript and have agreed to the final content.
Kenya National Commission for Science Technology and Innovation funded this study through grants NCST/5/003/3rdCALL/017 and NCST/5/003/3rd CALL/016 assigned to RMA and PNN respectively.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We wish to thank Nyotu Gitau, Sarah Kagotho, Rose Mbeya, Fridah Kariuki and Waruguru Wanjau all of Trek Science Limited for their administrative support and acknowledge Janet Irungu for her chemistry expertise and provision of equipment for HPLC and Hosea Mokaya for training in HPLC techniques. We also wish to acknowledge Chris Beadle for his innumerable revisions of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 14 Aug 17 |
|||
|
Version 1 03 Oct 16 |
read | read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)