Keywords
BSE, bioassay, 301V, sPMCA, in vitro, prion
BSE, bioassay, 301V, sPMCA, in vitro, prion
The transmissible spongiform encephalopathies (TSE or prion diseases) form a group of infectious and fatal neurodegenerative diseases affecting several species of mammals for which there is no available treatment or cure. The cause is thought to be a novel infectious agent (the prion), itself a misfolded isomer (PrPSc) of a benign cell associated protein known PrP or PrPC. This group of diseases include scrapie in sheep and goats, Creutzfeldt Jakob disease (CJD) in humans and bovine spongiform encephalopathy (BSE) in cattle. The UK BSE epizootic of the mid-1980s to early 1990s was the result of recycling BSE through the cattle food chain via a high protein feed additive known as meat and bone meal. It is thought that over 460,000 BSE infected UK cattle entered the human food chain before a ban on feeding specified risk materials to cattle came into force1. As a consequence of this, a new human disease referred to as vCJD, the human form of BSE, began to present in a number of young adults from the mid-1990s. This demonstration of the zoonotic potential of prion diseases generated an accelerated program of research into these diseases and much of this has required animal models. The murine passaged BSE strain known as 301V was first described by Bruce and colleagues2 during transmission studies of cattle BSE to wild type mice. BSE 301V is the product of serial passage within the VM mouse line and this combination of 301V/VM has been well characterised and used in numerous studies, including those aimed at understanding the fundamental brain pathology during neuropathogenesis3. In addition, the 301V/VM model has also been important in experiments analysing the effectiveness of various decontamination measures for BSE infectivity. For example, Taylor et al.4 demonstrated the effectiveness of formic acid in inactivating both 301V and scrapie in the context of occupational exposure to histological samples. This BSE model has also been used to show the lack of sufficient inactivation of BSE prions during historical rendering processes which resulted in the BSE outbreak in UK cattle5. For development of safe procedures in the context of human health, 301V has been used to model vCJD in the fractionation of plasma and the safe manufacture of blood products6, and additionally has been used to estimate BSE infectivity that is likely to remain after processes in the derivation of bone gelatine from bovine products7. A further study by McLeod et al.8 screened a number of different proteases for their ability to reduce the infectious titre of 301V as a novel method for the decontamination of sensitive surgical instruments. More recently, the availability of transgenic mice expressing the bovine PRNP transgene have become available, and with this their high susceptibility to bovine BSE prions has complemented the use of 301V in these types of experiments. A study published by Giles et al.9 directly compared the effectiveness of decontamination of both bovine BSE and 301V in transgenic and VM mice, respectively. 301V was more sensitive to both heat and chemical denaturation than cattle BSE, suggesting that the physical properties of the 301V BSE strain have diverged slightly from those of cattle BSE. Despite the more recent availability of these transgenic rodent strains the 301V/VM infection system remains a useful, well characterised model for BSE in TSE research and allows direct comparison with numerous previous studies.
In the last 15 years or so prion research has been revolutionised by the demonstration of in vitro assays that are thought to replicate the molecular events occurring in vivo during prion infection and the conversion of PrPC to the disease isomer PrPSc. First reported by Saborio and colleagues in 200110 the protein misfolding cyclic amplification (PMCA) assay is able to replicate prions in vitro within a source of PrPC (generally produced from a healthy brain homogenate) during cycles of PrPC to PrPSc seeded conversion followed by sonication with high frequency sound waves that break up aggregates of PrPSc to form new seeds or sites of nucleation. The products of this sensitive in vitro assay retain the biochemical characteristics of the prion seed and are infectious11. The sensitivity of the PMCA assay was improved by including the dilution of the reaction into fresh PrPC substrate after an optimal period of amplification. This modification, known as serial PMCA (sPMCA)12 has been widely adopted by the research community and has been applied to several rodent prion strains13, scrapie in sheep14,15, BSE in cattle16, and CWD of cervids17. sPMCA can achieve levels of sensitivity significantly beyond that of animal bioassay18 and these experiments take days or weeks to perform compared to the months to years of animal bioassay, and at a fraction of the cost. As such, amplification of prions by sPMCA can be used as a surrogate for measuring infectivity in vivo. To date, the 301V strain of BSE has not been used in sPMCA based studies. Here, we describe a high sensitivity 301V sPMCA that can, over a period of 5 days detect higher dilutions of infectivity than are attained by a 170–200 day bioassay within the VM mouse line.
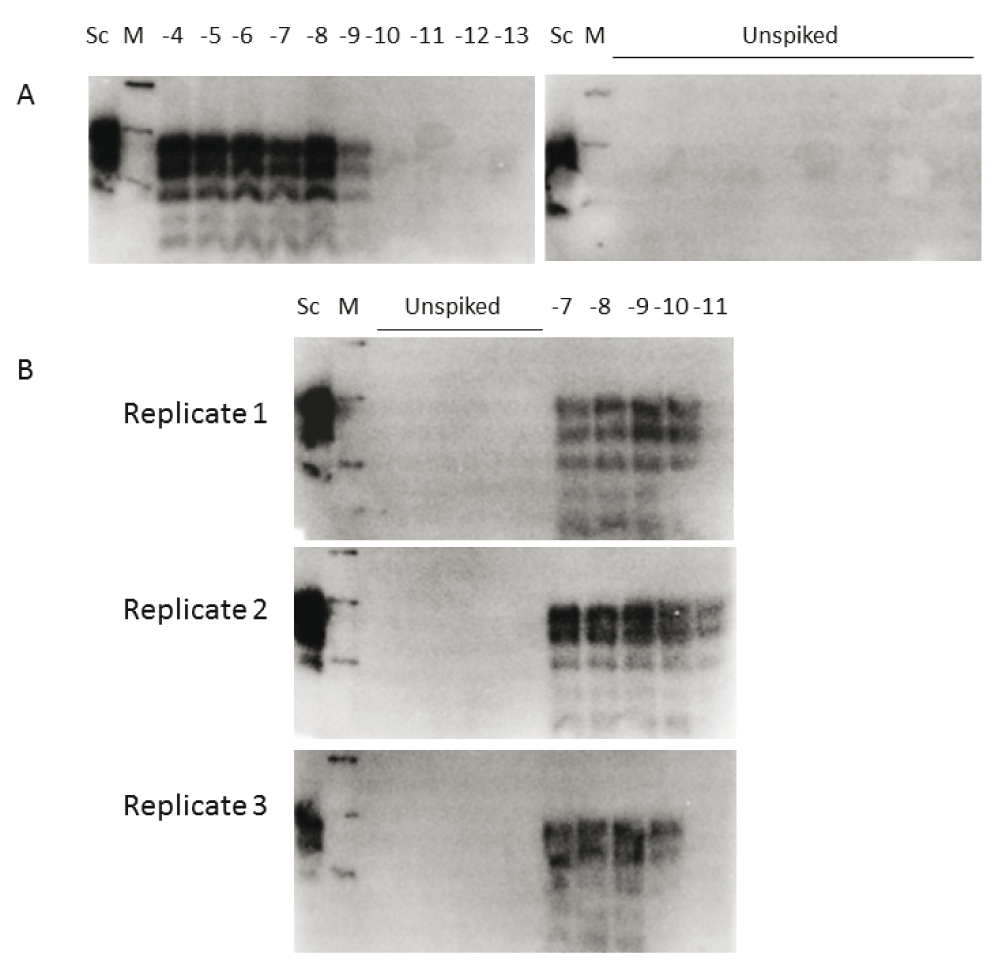
A. Reactions were seeded with 10 µls of 10-4 to 10-13 dilution of 301V brain (as indicated). Unspiked samples had 10 µl VM brain substrate only. Sc, scrapie positive brain sample was used as a blotting control. B. The assessment of reproducibility of the 301V sPMCA using three separate batches of VM substrate, each were seeded with 301V brain dilutions 10-7 to 10-11 (as indicated). Unspiked PMCA samples were always negative (a total of 15 replicates are shown). Western blots were probed with the anti-PrP antibody SHa31, M, molecular mass markers at 41, 30 and 22 kDa.
A pool of 301V mouse brain homogenate was used to assess the efficacy of a new sPMCA assay alongside conventional 301V/VM bioassay. For the in vitro assay we used a 5 day amplification method and a murine VM substrate. This 301V sPMCA assay demonstrated assay sensitivity to 1×10-9 dilution of brain homogenate (Figure 1A). The batch-to-batch variability test of a further 3 substrate preparations detected 301V to at least the same level (Figure 1B). The observed variations in sensitivity within these limiting dilution experiments (between 10-9 and 10-11) is likely a reflection of differences in individual substrate preparations that will be seen when making small volume preparations from limited numbers of brains.
The same 301V sample was also analysed in a VM mouse bioassay by limiting dilution and this bioassay detected infectivity in 1 out of 12 mice at the 10-8 dilution of 301V brain material (Table 1), equating to a 301V titre of the original brain pool of 108.5 LD50/g (as determined by Karber methodology19).
*Number of clinical and pathology positive mice/total injected. Mouse numbers exclude intercurrent deaths i.e. animals dying earlier than the 1st clinical case (there was one intercurrent death of a mouse receiving 10-7 brain dilution, and two receiving the 10-4 dilution). Total number of challenged mice were 6 per group for 10-4 and 10-5, 12 per group for 10-6 to 10-9.
| 301V Brain Dilution | No. mice positive* | Incubation period (days) | SD |
|---|---|---|---|
| 10-4 | 4/4 | 142 | 10 |
| 10-5 | 5/6 | 149 | 14 |
| 10-6 | 10/12 | 170 | 12 |
| 10-7 | 4/11 | 191 | 18 |
| 10-8 | 1/12 | 184 | - |
| 10-9 | 0/12 | - | - |
The 301V sPMCA assay can therefore detect PrPSc at a level at least tenfold more sensitive than the VM mouse bioassay, in a total assay time taking little over a week. Whilst we report sensitivity of the assay at 5 days of amplification, it is very likely that much higher levels of sensitivity could be attained with additional rounds of amplification. The highest dilution of 301V infectivity that could be detected within the VM bioassay was a 1×10-8 dilution of brain at 184 days post inoculation, or 26 times longer than the sPMCA assay. Maintaining animals within bioassay, including their category 3 containment make these kind of titration experiments very costly and time consuming to carry out. That, coupled with the ethical implications of use of animals means sPMCA could be the method of choice unless there is a good scientific reason for requiring to demonstrate infectivity (the ability to cause disease), or a requirement to monitor strain phenotype, as opposed to the surrogate marker of disease, PrPSc protein. A useful way of incorporating these two assays into future studies, could be to assess 301V seeding activity within a wide range of samples to identify those that contain PrPSc. Bioassay could then be used on a limited number of sPMCA-positive samples to confirm the presence of BSE infectivity. Another example of the application of sPMCA that has been routinely used for the detection of prions in a rodent prion model is with cervid CWD17. In this instance, CWD amplification within cervid CNS tissue substrate is notoriously inefficient, and transgenic mice have been used as an animal bioassay model for infectivity studies and also to provide substrate to facilitate efficient in vitro amplification by sPMCA.
In summary, we have developed a reliable in vitro method (sPMCA) for the detection of PrPSc resulting from infection with 301V (mouse passaged BSE). The assay is at least as sensitive as mouse bioassay and can derive data on the presence of PrPSc in a fraction of the time. This will be useful in studies such as those looking at BSE decontamination where the screening of large numbers of samples is required.
All use of animals, the collection of animal tissues and the use of such tissue was carried out in accordance with the Animal (Scientific Procedures) Act (ASPA) 1986, under licences from the UK Government Home Office (Project licence 60/2544). All animal experiments were subject to review and approval (01-124) by The Roslin Institute Ethical Review Committee and euthanasia methods were approved by the UK Home Office.
A serial dilution of pooled murine VM brains that were taken from 301V challenged animals was made as previously described20. A dilution series of this brain homogenate from 10-1 to 10-10 was made up in saline and used to inoculate groups of VM mice, bred in house at The Roslin Institute and of mixed sex, 6 weeks old (groups of 6 mice at 10-4 and 10-5, 12 mice from 10-6 to 10-10) with 20 µl of each dilution intracerebrally, as previously described20. Animals were observed daily for signs of ill health and euthanised by cervical dislocation when clinical signs of neurological disease or any intercurrent illness were observed. After euthanasia, brain tissue was confirmed as 301V positive or negative by detection of brain tissue vacuolation by light microscopy after Haematoxylin and Eosin staining20. This analysis was carried out blinded to the identity of the tissue in each case.
VM brains from healthy animals were supplied frozen, before preparation of the 10 % (w/v) homogenate substrate. Preparation of 10 % brain homogenates as substrates for sPMCA has been previously described21. Here, we included the sPMCA additive digitonin22 (Sigma-Aldrich) which was added to reactions at 50 μg/ml. sPMCA reactions were assembled in 200 µl thin wall PCR tubes, and comprised 90 µl brain homogenate substrate with digitonin, three 2.4 mm Teflon beads (Precision plastic ball co. Ltd) and 10 µl of 301V sample to be amplified (10-4 to 10-13 dilution of 301V brain). Unspiked negative control samples were set up substituting the 301V seed with 10 µl VM brain substrate only. Reaction tubes were placed in a Misonix S3000 sonicating water bath set on a program of 10 seconds sonication every 30 mins, for 24 hours at a power setting of 190–200 W at 37°C. Every 24 hours, samples were diluted 1 in 10 into fresh VM brain substrate and sonicated for a further 24 hour round of repeated sonication and incubation retaining the same three Teflon beads throughout the 5 rounds of sPMCA. Amplifications were carried out for a total of 5 days. Dilutions of 301V brain homogenate are recorded as the dilution of brain spike before addition to the amplification reaction, ie 10-1 is 10 µl of a 10 % (w/v) preparation of brain, 10-2 is 10 µl of a 1 in 10 dilution of the 10-1 preparation of brain etc. All sPMCA was carried out at 37°C in a Misonix S3000 microplate horn.
sPMCA reaction products (10 µl) were digested using a final concentration of 50 µg/ml proteinase K (Sigma-Aldrich), for 90 minutes at 40°C. Samples were then boiled for 5 minutes in 1X LDS buffer (Invitrogen) and electrophoresed through a NuPAGE SDS-PAGE gel system (Invitrogen) using 12% (w/v) acrylamide gels. Molecular mass markers (prestained Seeblue plus2, Invitrogen LC5925) were run alongside samples. As a blotting control an aliquot of proteinase K digested (50 µg/ml proteinase K (Sigma-Aldrich), 60 minutes at 40°C) scrapie positive ovine brain (equivalent to 2 µl of a 10% w/v brain homogenate) was also loaded onto each SDS-PAGE gel. Proteins were transferred to PVDF (Roche) membrane by electroblotting, and the membranes were then blocked for 1 hour with 3 % (w/v) skimmed milk. Western blots were probed with the anti-PrP mouse monoclonal antibody SHa31 (SpiBio A03213) diluted to 1/80,000 and a polyclonal goat anti-mouse immunoglobulins Horse Radish Peroxidase conjugate (Dako P04477), diluted to 1:20,000, as previously described21. Blots were imaged after the addition of EZ-ECL HRP substrate (Geneflow) using an ICCD225 photon counting camera system and IFS32 image software (Photek Ltd).
F1000Research: Dataset 1. Raw uncropped images of the Western blots shown in Figure 1. 10.5256/f1000research.9735.d13863823
RAS and NH conceived the study and provided both the 301V and VM biological material, BCM and KCG devised and directed the experiments, KB carried out the experiments. BCM wrote the manuscript, all authors proof read the manuscript before submission.
This work was funded by DEFRA under project SE1433 (Robert Somerville) and by BBSRC Institute Strategic Grant BB/J004332/1 (The Roslin Institute).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
References
1. Boerner S, Wagenführ K, Daus ML, Thomzig A, et al.: Towards further reduction and replacement of animal bioassays in prion research by cell and protein misfolding cyclic amplification assays.Lab Anim. 2013; 47 (2): 106-15 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
References
1. Faburay B, Tark D, Kanthasamy AG, Richt JA: In vitro amplification of scrapie and chronic wasting disease PrP(res) using baculovirus-expressed recombinant PrP as substrate.Prion. 2014; 8 (6): 393-403 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 18 Oct 16 |
read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)