Keywords
Hyperspectral, Sonification, Fluorescence, Microscopy, Visualization, Multiplexing, FIJI, ImageJ
Hyperspectral, Sonification, Fluorescence, Microscopy, Visualization, Multiplexing, FIJI, ImageJ
The increased availability of microscopes with multiple spectral channels and multi-colored fluorescent molecular markers has allowed life science researchers to generate datasets of higher spectral complexity than ever before from their optical imaging systems1. By selectively placing different markers throughout a specimen, a researcher can construct detailed visual narratives of different biological processes based on the spectral variations throughout the data2–5. This approach, called multiplexing, allows for functional readout of multiple genetic functions spatially and temporally, a feature unique to fluorescence imaging. These multiple fluorophores can be genetically engineered extrinsic fluorescence tags such as Green Fluorescent Protein (GFP) or endogenous fluorescence such as that emanating from cellular autofluorescence. In particular, spectral information has been used to discriminate fluorescence signals from cellular autofluorescence signals such as nicotinamide adenine dinucleotide (NADH) in metabolism studies and to understand subcellular protein trafficking via signaling protein labeling6,7. Additionally the collection of spectral data enables researchers to study other complex phenomena related to fluorescence such as fluorescence resonance energy transfer (FRET)8 or spectral shifts.
As fluorescence microscopy datasets increase in richness and dimensional complexity, designing flexible tools for researchers to explore these larger and denser datasets that strike the appropriate balance between intuitive functionality and analytic effectiveness is becoming increasingly important.
As the number of fluorescent markers used in a biological sample increases, two critical bottlenecks emerge within the current traditional analysis workflow. First, the spectral properties of fluorescent dyes often necessitate a linear unmixing (deconvolution) of the image data to discriminate one dye from the other. The importance of this step becomes particularly apparent when simultaneously using many fluorophores with overlapping emission spectra. Several types of spectral stripping and unmixing techniques have been borrowed from other scientific domains such as space science that involve analysis of hyperspectral data9,10. However, many of these techniques require precise a priori knowledge of the present spectra before the imaging5,11–13. Confirming emission spectra in experiments involving biological specimens becomes complicated due to factors such as spectral variance in endogenous chemical environment, the developmental fluorophore stage, and unpredictable refractive characteristics inside the specimens. All can cause significant variation in emission spectra14.
Second, even if all spectra are successfully unmixed, visual representations of an image volume with more than three spectral channels are limited by inability of the human vision system to manage effectively more than three channels at a time. False color images of datasets with three or more spectral dimensions can visually appear to be the same in areas where the data are vastly different, because the transcoding operation used to render the raw data visible to human eyes involves a data loss. Therefore visual analysis of hyperspectral imagery has to always rely on analysis of multiple images or on custom computational pre-processing.
While the human vision system is limited for natively comprehending rich hyperspectral data, the human ear and auditory system are more optimally suited for this task15. Figure 1 shows a spectral sensitivity curve for the human eye, while Figure 2 shows the “equal loudness curve” describing the range of frequencies perceptible by the human ear. Note the logarithmic scaling on the x-axis of Figure 1 as compared to the relatively narrow interval on the x-axis of Figure 2. Additionally, whereas the eye has only three channels of spectral sensitivity, the ear can distinguish about 1400 pitches throughout its range16. Moreover, the ear actually has finer resolution when sensing complex, non-sinusoidal tones15. This behavior makes the ear a substantially more optimal sensor for spectrally rich signals than the eye, because, as we demonstrate, in Figure 1 and Figure 2, the eye actually becomes less spectrally adept with increasing spectral complexity. We intend to show how sonification allows researchers to avoid complex computational and visual techniques to best interpret their hyperspectral datasets.

This chart shows the combined spectral sensitivity of all the color cones in the human eye, which constitutes the full range of visually perceptible frequencies.

This chart shows the spectral sensitivity of the human ear in the form of multiple equal loudness contours over a frequency domain. While the eye has a domain of several hundred Hz, the ear has domain spanning several thousand Hz, as seen by the log scaling.
Research on data sonification has been conducted across a diverse variety of disciplines and has yielded results across the spectrum from purely aesthetic to highly functional. The emphasis in our project was on creating a sonification plugin as a functional and practical tool for fluorescence microscopy research. As such, we decided to design our sonification as an extension of the imaging and analysis workflow that researchers at our lab use to gather and understand multispectral fluorescence microscopy data. Sonification has been proven a viable means for analyzing hyperspectral or otherwise dimensionally complex datasets in many other contexts17–19. Potential microscopy sonification applications have been briefly explored but not actively pursued and implemented by the scientific community20,21. Dr. Thomas Hermann, in his 2002 PhD thesis “Sonification and Exploratory Data Analysis”, considers the viability of a hypothetical sonification-based multichannel image analysis tool for microscopy research22. In his thought experiment he outlines three ways that sonification would uniquely provide additional clarity and insight into dimensionally complex microscopy datasets:
--Sonification intuitively allows users to perceive trend-related factors such as clustering, or other distributional behavior with increased clarity by enunciating more subtle spectral differences that missed by more conventional data display methods. This allows users to intuitively understand higher order relationships amongst the 15+ channels that might go unnoticed when visually exploring the image stack 1–3 channels at a time.
--Sonification augments perception when the eyes are saturated with other information. If a user needs a visual interface to explore the structural organization of a particularly intricate specimen, the sonification can simultaneously provide information on salient characteristics from other channels co-localized to a particular region of interest without obfuscating the visual-spatial information.
--Sonification enhances the salience of 'warning flags'. Rapid changes in sound naturally command the human attention very effectively. It is a lot easier to grasp the difference in amplitude between two points in the dataset than it is to compare the intensity of two points defined by multiple overlaid color maps.
Currently, no data sonification package exists for hyperspectral image volume exploration and analysis that avoids computational dimensionality reduction. Researchers at Stanford have implemented a sonification model for hyperspectral image analysis, but this relies heavily on principal component analysis and avoids sonification of the raw multichannel pixel stream21. Our approach uses data sonification to reduce the amount of abstraction between the researcher and the raw fluorescence data. We explore sonification as a way of confirming a spectral deconvolution, as well as a way of navigating the fluorescence data in its full dimensionality. By allowing researchers to interact directly with the spectral composition of their datasets, we can potentially elucidate salient characteristics of the data that otherwise would have gone unnoticed.
There are two components to the sonification. The first is the image-viewing interface that provides the user interface for the data to be explored and sonified. For this we chose a plugin context inside FIJI (http://fiji.sc/) which is a popular distribution of the Java-based image analysis program ImageJ23,24. Choosing an open-source image analysis package was important to us as to allow maximum accessibility and ease adoption. FIJI is already the tool of choice for many scientists doing microscopy research. Several of the data sonification implementations described in the current research literature use a standalone application which integrates image UI and audio driver communication into one executable, but this was not the optimal strategy for our proposed use case. A FIJI plugin was the intuitive choice for development and deployment because it can easily be integrated into current research workflows instead of requiring the installation and usage of a separate standalone application. Additionally, since FIJI is used for many spectral fluorescence microscopy analysis routines, it opens up the possibility of integrating and interacting with these steps as needed by the end user.
The second component is the audio server, which is tasked with storing synths and outputting sounds to the native audio driver. Synths are definitions that describe the range of sounds produced by the sonification, and contain instructions on how to translate the commands received from the image host into audio samples. As we discuss in the “Sound Design” section, iteratively designing multiple mappings from data-space to sound space is how we explore the capacity of sonification to allow us to interact with hyperspectral datasets. We decided to use SuperCollider (http://supercollider.sourceforge.net/) and JCollider (http://www.sciss.de/jcollider/) for the functionality of storing synths and communicating with the audio driver. SuperCollider is an open-source package for algorithmic sound synthesis, and JCollider is a library that allows Java applications, such as ImageJ, to communicate with an instance of SuperCollider’s OpenSoundControl (OSC) server, called “scsynth”. This communication entails encoding and sending audio commands, manipulating synth definitions, and checking server status. Synth definitions are encapsulated in SynthDef Objects, as specified in the JCollider API, and exported to scsynth at runtime. All communications between the Java image host and the OSC server are done over User datagram Protocol (UDP).
The following signal flow diagram in Figure 3 describes the interaction process of the plugin. The user interacts with the image via the FIJI interface by clicking and dragging the cursor across the image, which sends pixel data encoded in OSC format to scsynth. The data are then sonified by the SuperCollider server in accordance with the chosen synth definition and sent to the speakers as audio samples.
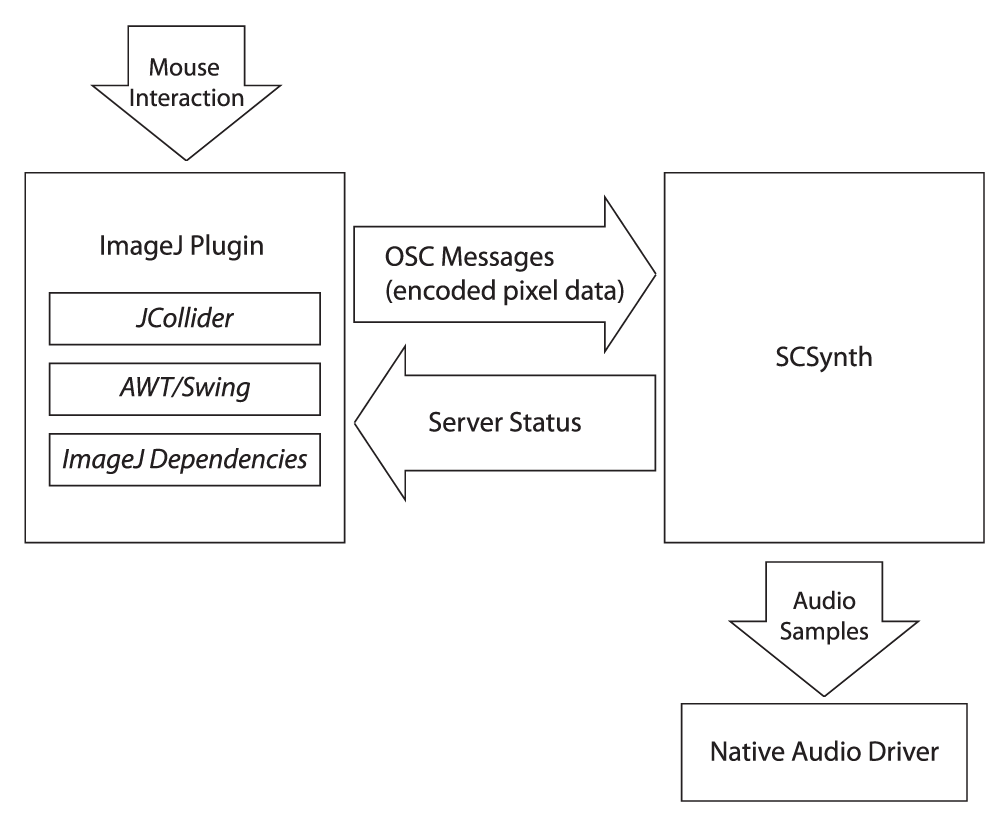
This signal flow diagram describes the data sonification pipeline. When the user interacts with the image volume by clicking and dragging, FIJI sends encoded pixel values to the SuperCollider server, which translates the data into audio samples sent to the speakers.
When designing the mapping from data space to sound, we first had to establish a set of heuristics to determine what qualities of the sound are desirable in a particular mapping. Grond and Berger’s chapter in the “Sonification Handbook”, entitled “Parameter Mapping Sonification”, informed our sound design methodology25. They outline the following optimal design criteria for a parameter based exploratory sonification:
Polarity. Polarity constitutes the capacity of a sound to communicate both the direction and magnitude of a change in a stream of data. In the use-case of sonifying a spectrally rich signal, as an individual component of the set of spectra increases or decreases, such as the pixel intensity value for a particular channel in an image volume, the respective mapped quantity of the sound should change in a way that can be clearly identified as the increase or decrease.
Scale. Data from our image volumes have to be appropriately scaled in the mapping process in order to convey the full range of parameter fluctuations. Ideally the same image in two different formats should sound the same, so normalization of the pixel stream data should be implemented to minimize arbitrary differences in auditory characteristics due to image encoding and prevent signal spikes. Images encoded in a format with higher dynamic range have higher amounts of noise though, so there will invariably be some differences.
Context. Context is the auditory equivalent of “axes and tick marks”--elements of the sonification that provide a baseline reference against fluctuations in the data25. If fluctuations appear to be arbitrary or ambiguously connected to observed changes in the data, the utility of the sonification as an exploratory mechanism is severely diminished. As such, we sought to design our synths with as little auditory clutter as possible; one should be able to selectively focus on particular modulating qualities in the sound and intuitively know how the data are changing. This rule of thumb definitely informed our preference for additive synthesis. Another issue related to context we observed was that, similar to the case with color lookup tables, there is no “one size fits all” auditory display solution. Certain synth designs are more useful for articulating subtle changes in spectral composition but ineffective for perceiving larger trends across the entire dataset, and vice versa. As such, we provide a variety of synths, each for usage in a specific observed context.
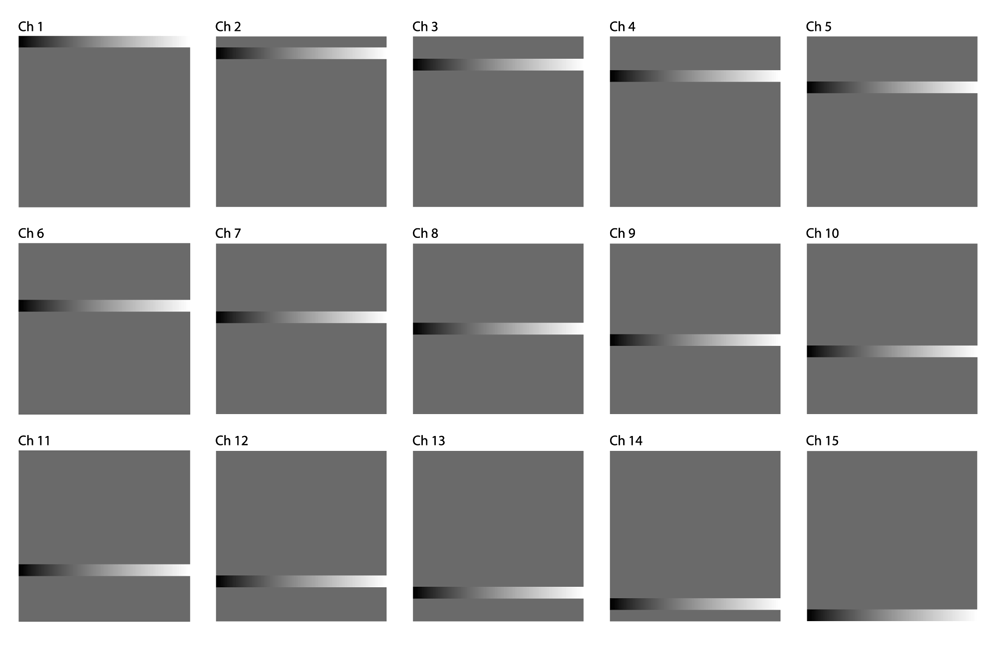
Calibration. We employ a series of calibration images designed to systematically screen any arbitrary synthesis algorithm for viability and identify what spectral contexts are or are not displayed clearly by a given sound. Figure 4 shows a comparison of the four volumes we generated. Each calibration image volume consists of 15 450px by 450px TIFF images representing the spectral data shown in Figure 5a–Figure 5d.
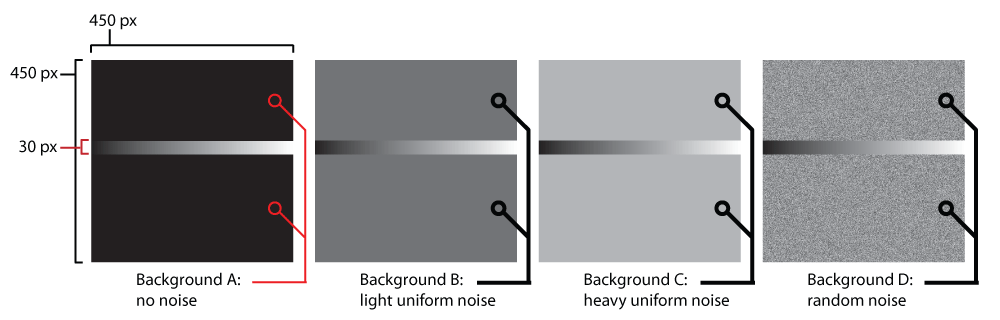
Single channel slices of each of the four calibration image volumes are compared side by side. In each volume, the same cascading gradient is interpolated with a different ‘background noise’ texture. This particular design of a calibration image allows for testing the signal-noise discrimination capabilities of a given synth for each channel in a variety of spectral contexts.

Calibration Volume ‘A’ contains the cascading 30px gradient with no background noise. This is a control volume to test a particular synth’s range of sounds with no background interference.
Calibration Volume ‘B’ contains the cascading 30px gradient with low background noise. This volume tests a particular synth’s signal-noise discrimination with low intensity homogeneous background noise across all channels.

Calibration Volume ‘C’ contains the cascading 30px gradient with high background noise. This volume tests a particular synth’s signal-noise discrimination with high intensity homogeneous background noise across all channels.
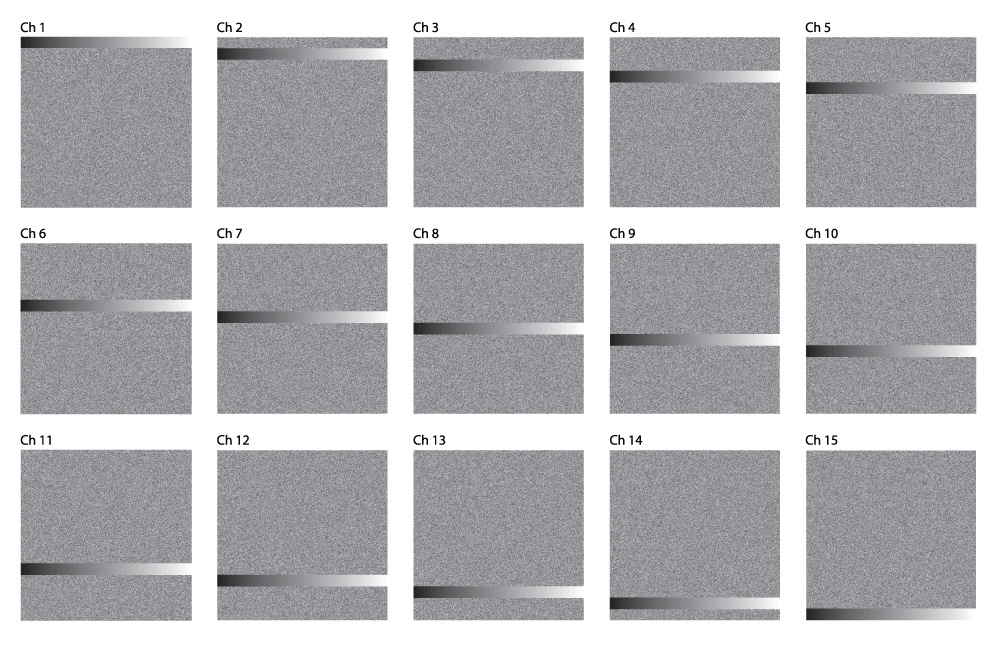
Calibration Volume ‘D’ contains the cascading 30px gradient with random background noise. This volume tests a particular synth’s signal-noise discrimination with pseudo-random, heterogeneous background noise across all channels.
Note in Figure 4 there is a 30 pixel thick horizontal stripe that traverses the entire width of the image in each of the volumes. This stripe starts at the top of the image in channel 1 of each volume, and incrementally shifts downward 30 pixels each channel until it is at the very bottom of the image in channel 15, as seen in Figure 5a–Figure 5d. Each of these stripes has gradation from 8-bit grayscale intensity value 0 (black) on the leftmost side to 255 (white) on the rightmost side. Throughout the rest of the image not occupied by the stripe, each of the four calibration volumes has a different pattern, designed to simulate various types of noise that could interfere with the perception of the 0–255 gradient. Volume A is designed to be the control volume which should showcase the discriminatory function of a sonic mapping at its clearest. Volumes B and C both have noise that is homogenous throughout the entire background; B has light noise (grayscale value = 84/33% intensity) while C has heavier noise (grayscale value = 168/66% intensity). Volume D has pseudorandom noise that is clamped between 20% and 80% intensity (to prevent unnecessary clipping).
When the 15 images of a calibration volume are superimposed on top of one another in an image stack, the cascading stripe and background pattern provides a means to measure the effectiveness of a particular sonic mapping at discriminating intensities for any given channel by traversing the image horizontally. Conversely, the spectral differentiation of the mapping can be gauged at a given intensity by traversing the image vertically. Movie 1 shows how sonifying these four volumes with a given sound can give an idea of its effectiveness in a variety of spectral contexts.
Each synth definition we created is outlined here as a list of summations defining how audio samples. These following synths were utilized on 15 channel datasets, so there are 15 components to each synth waveform that are modulated by fluctuations in the spectral content of a given stream of pixels. Additive synthesis is the primary algorithm used to group all these fluctuations into one sound in the definitions we created.
Synth 1.
f = {440Hz, 466.16Hz, 493.88Hz, 523.25Hz, 554.37Hz,
587.33Hz, 622.25Hz, 659.26Hz, 698.46Hz, 739.99Hz,
783.99Hz, 830.61Hz, 880Hz, 932.33Hz, 987.77Hz}
ia = normalized pixel intensity value (0–1) for channel ‘a’ of a multichannel image volume
This synth maps the intensities of spectral channels 1–15 to the amplitudes of sine waves from A4 to B5, and then adds all the waves together to produce the resultant sonification for a given pixel. We found that this synth design was effective for the phantom, but was less useful for articulating subtle spectral distinctions in biological datasets.
Synth 2.
This synth uses the same additive principle of Synth 1 (intensities mapped directly to amplitude), but for a different set of notes. Instead of incremented tones on a scale, this synth uses a harmonic series. We found this synth to be notably more useful when sonifying biological datasets than Synth 1 as the wider range of notes makes it more difficult for the user to mistake adjacent spectral channels for one another.
Synth 3.
This mapping is also an additive synthesis algorithm, but instead of modulating the amplitudes of the components, the actual frequencies are modulated to create inharmonics. The amplitude of each component is set to 0.1 (statically), and the intensities of spectral channels 1–15 are mapped to a range of 0–150Hz deviation from each harmonic partial.
Synth 4.
This mapping combines modulation of amplitude with deviation from harmonic partials. We found the combination of the two allowed for better spectral differentiation than either Synth 1 or Synth 3 alone.
Synth 5.
This is the mapping we used in all the example videos. The summation is constructed the same way as Synth #4, except the range of notes is expanded by choosing every other higher order harmonic partial of 300Hz. Additionally the frequency deviation from the harmonics, mapped to the pixel intensity, is scaled to 350Hz as opposed to just 150Hz. We found it to display subtle spectral changes with the most clarity out of all the synths.
The human eye makes use of photoreceptors with overlapping spectral sensitivity in order to improve its spectral resolution. It is important to note that this method does not increase the total number of colors or color combinations the eye can perceive, but rather optimizes color perception in certain situations that are more likely to be relevant in a natural environment while decreasing resolution in other situations. Specifically, overlapping color receptors allow the eye to establish the mean frequency of a single emission peak with greater accuracy. When, however, multiple peaks are involved this method becomes unreliable since there are always different peak configurations that will lead to the same perceived color. To illustrate this effect below we present a simple method to generate those ambiguous peak configurations for any given photoreceptor sensitivities. We demonstrate this method by generating a dataset made up of five emitters. We produce a hyperspectral image in which every pixel has a different spectrum due to different relative fluorophore concentration or emission wavelengths, but all appear identical to a three channel detector with overlapping spectra such as the eye.
Consider a sensor with three overlapping channels detecting a spectrum consisting of five relevant channels. The input spectrum can be described as a five-element vector I and the detected intensities at the sensor are described by a three-element vector D. D can be computed from I by multiplying a 3 by 5 matrix M that in its columns contains the spectral sensitivity curves of the three spectral channels.
We consider D and M fixed and are seeking all vectors I that fulfill the equation
D = MI
By varying u and v we can create a set of five channel spectra that all result in the same vector D. This D can contain negative numbers, which is not realistic. To avoid this we add set of constants Δ1, Δ2, Δ3, Δ4, Δ5 to our spectra I such that the smallest value occurring is zero. This leads to a different detected spectrum D’ that is still constant for all incident fluorophore spectra:
The result of this procedure, our 5-channel hyperspectral phantom dataset, is shown in Figure 6. Each pixel in the image multichannel volume has a different 5-fluorophore combination. They all look identical to the three channels sensor used. The spectra do however sound dramatically different in a simple sonification.
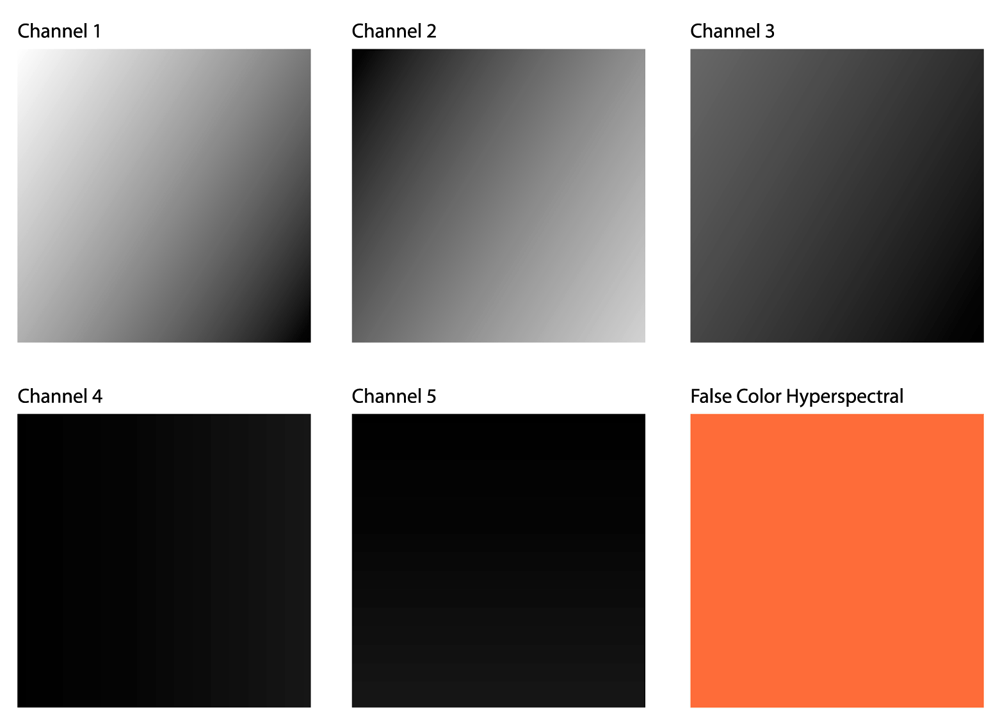
This figure shows a visual comparison of each channel of the phantom image volume alongside a hyperspectral false color image of the volume. While each channel has a unique gradient across the image, the hyperspectral false color image appears homogeneously textured throughout.
The sixth image seen in Figure 6 is a hyperspectral false color image of our phantom image volume. This false color image is a mapping of the 5D data to 3D space, using the spectral sensitivity curve seen in Figure 7, which has a spectral overlap similar to the cone sensitivity of the human eye shown earlier in Figure 1.
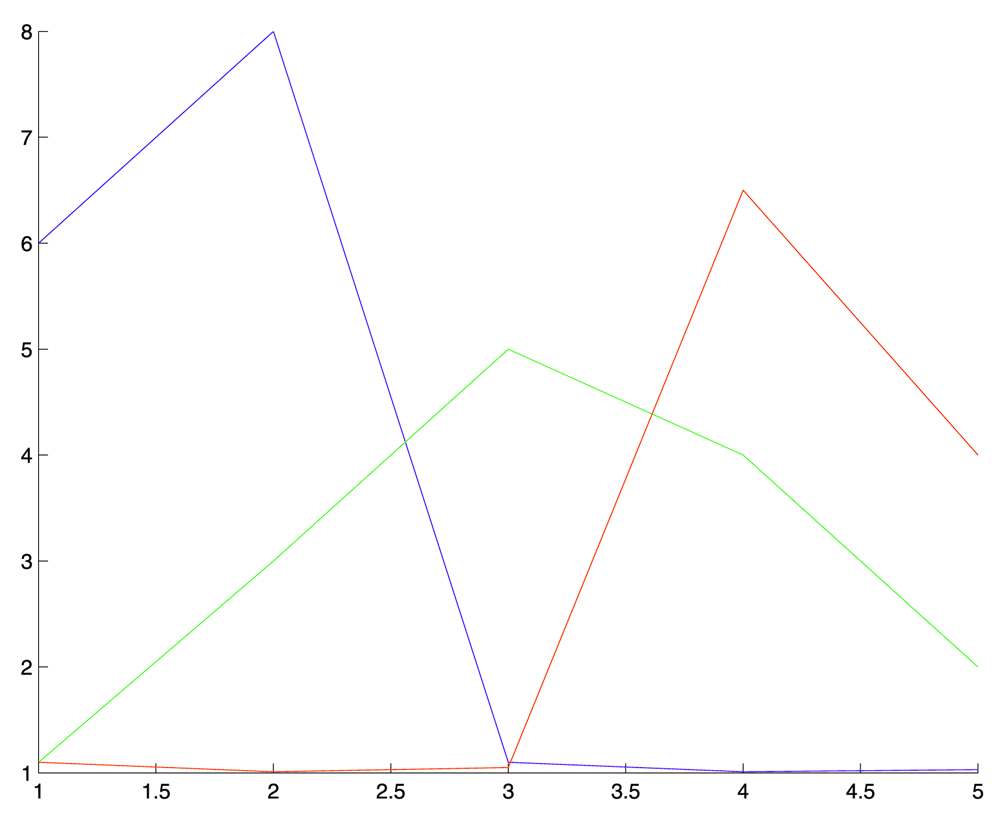
This figure shows the spectral sensitivity curve used to generate the hyperspectral false color image from the phantom image volume. In this curve the three color components are overlapping to emulate the sensitivity curve shown in Figure 1.
Although the false color image appears to be uniform across its entire surface, the actual spectral data, shown on the left, are different for every pixel. Even though there is significant overlap between the spectral peaks used in the false color mapping, there are no apparent visual distinctions in the RGB color image. Figure 8 shows the invisible variations in spectral content that characterize our image volume.
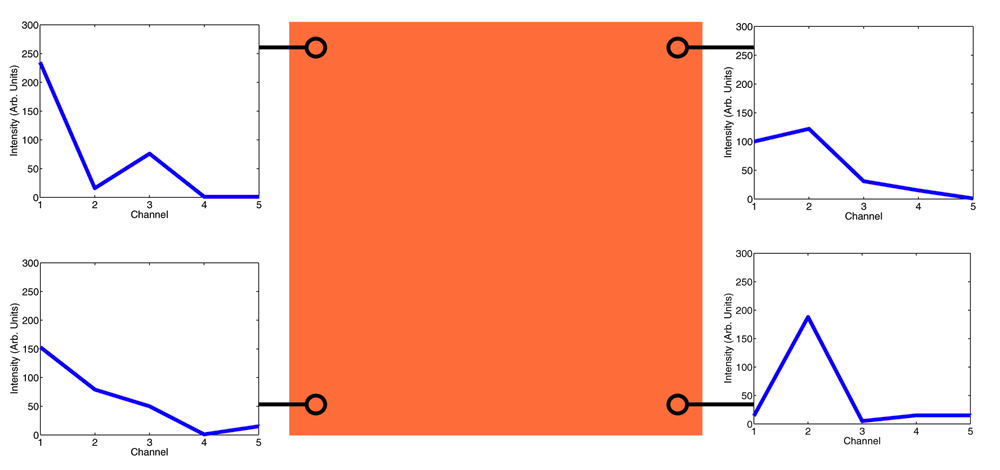
The spectral composition of the phantom image volume is shown at various points throughout the volume and overlaid with the hyperspectral false color image. This shows the range of spectral variations that are not visible through the false color image.
This data loss does not reflect an inadequacy in the sensitivity curve, but rather the fundamental inability of a three-dimensional transcoding to convey fluctuations in a five-dimensional signal. In the above section detailing the generation of the phantom dataset we describe how any arbitrary spectral response curve with three peaks can be used to compute a five-dimensional image volume that varies spectrally at each pixel, and a RGB false color hyperspectral image that appears to be spectrally homogeneous across all pixels to the human eye.
Using the sonification plugin, the user traverses the image volume and generates an audio signal that elucidates the invisible spectral topology of this image volume. A log on the left displays spectral data for each pixel that is sonified, confirming the spectral difference. This process is shown in Movie 2.
After demonstrating theoretical utility with the phantom dataset, we seek to create a test for our sounds that would more closely resemble the spectral subtleties of a real biological specimen with multiple fluorophores. However, we want a dataset that is much more spectrally controlled than a biological specimen so we can unequivocally verify our plugin’s functionality.
The FocalCheck DoubleGreen Fluorescent Microspheres Kit (Life Technologies, Carlsbad CA) provides a useful litmus test for using the sonification to differentiate two objects with very similar spectra. The three different fluorescent beads in this kit are uniformly labeled and are designed to test the spectral separation capabilities of a scope system. The subtle distinction in dye coverage and color between the three beads is designed to be visually unnoticeable, but precisely pronounced enough to be resolved by algorithmic linear unmixing. By testing our sonification with the FocalCheck beads, we can at once ascertain the capability of our plugin at confirming the results of a linear unmixing as well as differentiating spectra that are visually indistinguishable.
Figure 9 shows magnified false color images of the three beads with spectral measurements taken both at the core and the edge for each bead. The bead in Figure 9a homogeneously emits “green 1”(512 nm) and the bead in Figure 9b homogeneously emits “green 2”(525 nm). The third bead in Figure 9c is green 1 on the outside ring and green 2 in the center. Despite the spectral differences, all three false color images appear to have the same color.

The spectral content of the FocalCheck DoubleGreen bead 1 (512nm emission) is shown at the core and at the edge of the bead.
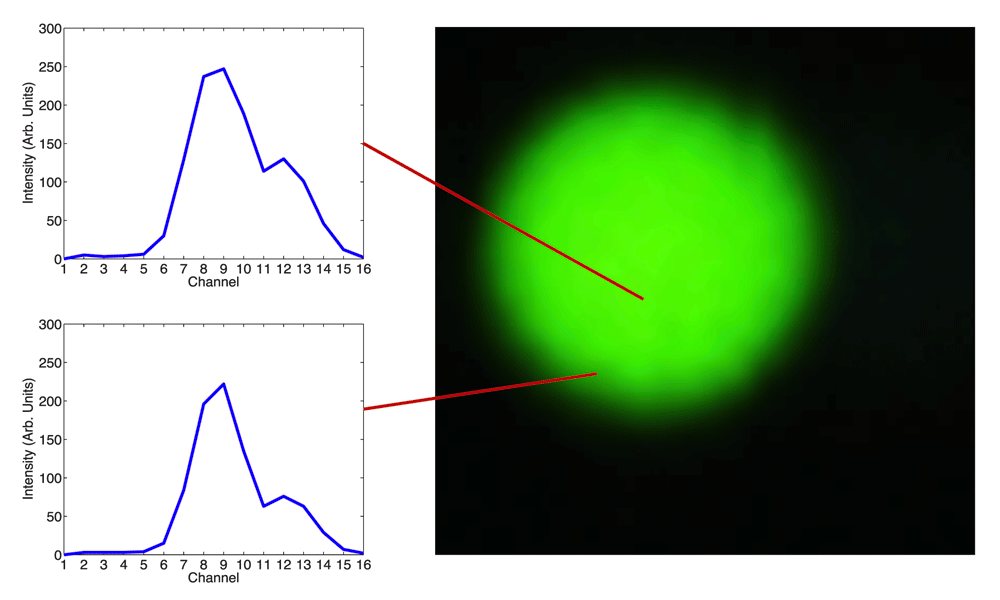
The spectral content of the FocalCheck DoubleGreen bead 2 (525nm emission) is shown at the core and at the edge of the bead.

The spectral content of the FocalCheck DoubleGreen bead 3 (512nm emission at the edge, 525nm emission at the core) is shown at the core and at the edge of the bead.

This figure shows a hyperspectral false color image of an Arabidopsis thaliana specimen with spectral measurements of the raw multichannel image volume taken at various points of interest. The spectral differences between the cell membrane and the plastid are obscured in the hyperspectral image.
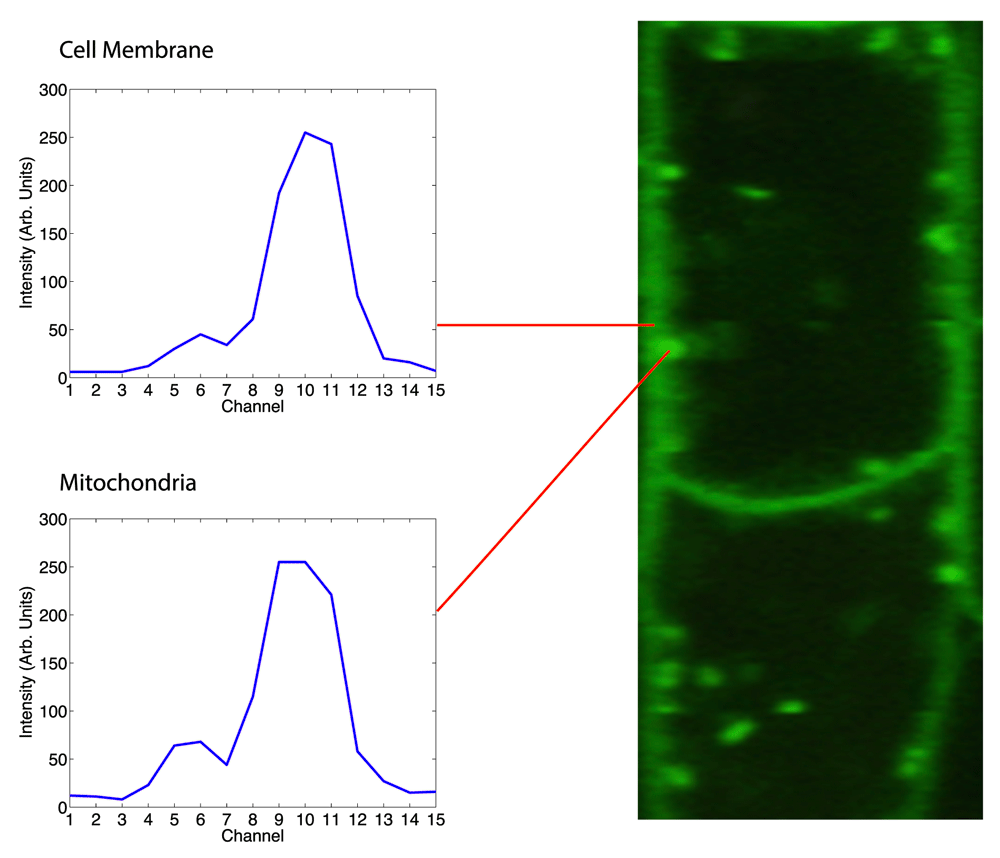
This figure shows a zoomed in hyperspectral false color image of an Arabidopsis Thaliana specimen with spectral measurements of the raw multichannel image volume taken at a cell membrane and a mitochondria. This presents another subtle spectral distinction in a live specimen that is not made clear by the hyperspectral image.
The video in Movie 3 shows a sonification of the same three image volumes. The cursor is clicked and dragged over each volume along a path that includes the blank space outside the bead, the edge of the bead, and the core of the bead. Choosing this path for each volume allows us to distinguish the “edge effect” of increasing intensity between the blank space and the bead (which is a function of scope resolution) from the subtle spectral shift between the thin ring around the outermost point of the bead and the bead’s core.
The final test of our sonification was sonifying a real biological specimen that has been modified to express multiple fluorescent proteins. We imaged an Arabidopsis seedling sample that expressed three fluorescent proteins, RFP, YFP and GFP, which have been genetically modified in accordance to the procedure described by Kato et al26.
In our first figure, we explore a visible similarity in the hyperspectral false color image between the plastids and the cell membranes of the Arabidopsis root. Although the plastids express a red fluorescent protein, background noise (likely from autofluorescence and scatter) in the green wavelength causes the plastids to appear very visually similar to the regions of the specimen expressing GFP.
However, when sonified, we can hear a very salient difference between plastids and the cellular membranes. Additionally, the plastids that are visibly red sound similar to the plastids with more green/yellow tinting, which allows the user to clearly understand the distribution of the plastids throughout the specimen despite the counterintuitive shift in color.
Next, we zoomed into an area of the specimen that has both mitochondria and cellular walls. The mitochondria express a protein (YFP) that is spectrally very similar to the protein the cell walls (GFP). In the false color image, this distinction is not apparent at all, due to the close proximity of the two spectral peaks.
The sonification clearly articulates the difference between the two peaks, even at varying intensities. In this video a user sonifies several parts of the image where mitochondria are either very close to or overlapping the cellular boundary, and the spectral difference is still made apparent.
Moving forward, the sonification framework that we have developed can be extended to provide deeper insights into complex microscopy datasets that have been inadequately represented through current visualization-only analysis methods. In particular, it is worth considering the potential utility of sonification-based analysis in experiments that involve multiplexed functional markers, such as calcium signaling, or experiments that require differentiation between intrinsic and extrinsic fluorescence. Additionally, improvements to the functionality of our plugin would allow sonification based analysis to be utilized in more demanding contexts. Ideas for improvements to the plugin fall under two main categories: improvements to the sonification interface, and extension of the proof of concept.
Improving the sonification interface could be done on multiple levels that would amplify the utility of the plugin. First, adding functionality such as sonification over a Region of Interest, or automatic exporting of a sonification to a video format would afford the plugin a higher degree of similarity to traditional image analysis plugins, making it more usable to scientists already familiar with FIJI. On a deeper level, an external GUI could be designed to allow for full programmability of the sonification mapping process. Similar to the way that Digital Audio Workstations provide virtual instrument interfaces for programming custom sounds through a synthesis algorithm, our plugin could benefit from a visual interface that would allow users to tweak the mapping parameters connecting data space to sound space to create the perfect sonification for a given use case. This would be the audio equivalent of creating a custom lookup table for image analysis. Combined with the calibration image volume and the documentation of our incremental sound design process we have provided, users would be able to use such an interface to intuitively find the most useful sounds for given datasets.
Extending this proof of concept application into new use cases would also expand the functionality of our plugin. We demonstrate how sonification can be used to confirm a given computational spectral separation, and as such it would be worth considering the utility of sonification alongside other computational image analysis techniques utilized on spectrally dense datasets. Moreover, it is worth considering the utility of sonification as the size and complexity of datasets scale, and whether parallel computing could potentially be useful for generation of sounds in real time with large numbers of input parameters. Finally, we have described ways that sonification provides us insights into our particular datasets that were inaccessible with only visualization, but it would be worth exploring in the general sense whether a difference can be quantified between a combination of sonification and visualization based analysis versus just visualization. Conducting an A/B test with a population of users and evaluating the variation in experience would be a good way to perceive the difference sonification makes in an applied context.
In this paper we have provided an in depth study of the utility of displaying spectral relationships in high-dimensional microscopy datasets. This has been the first study of sonification in microscopy without dimensionality reduction. Sonification has a wide variety of applications in image analysis, including providing data display to visually impaired users, clearly conveying patterns and relationships in complex data, and providing a novel way to aesthetically represent a dataset. As multiple fluorescent markers are becoming more important and common in biological microscopy research and interpretation of the resulting rich spectral data become more relevant, scientists will have to turn to new types of tools to meaningfully interact with their data. Sonification has potential to not only be used as an alternative to conventional spectral visualization but as well to complement in certain use cases where sonification would be especially advantageous. We have presented sonification as more than just an aesthetic novelty, but as a promising solution to address limitations in existing spectral visualization based image analysis techniques. Future work will need to further explore the practically of sonification of microscopy spectra in terms of appropriate sound mapping, biological application and user interface.
F1000Research: Dataset 1. Raw data for ‘Sonification of hyperspectral fluorescence microscopy datasets’, Mysore et al. 2016, 10.5256/f1000research.9233.d13868227
Latest source code for the sonification plugin: https://github.com/uw-loci/sonification
Archived source code at the time of publication: 10.5281/zenodo.15996028
The plugin can be installed through the FIJI Updater by following the LOCI updates website (http://loci.wisc.edu/software/fiji-plugins). The calibration images utilized to test our synth designs are available on the LOCI GitHub alongside the plugin source code, so as to facilitate the development of new data-to-sound mappings by users.
Figshare: Video of Calibration. doi: 10.6084/m9.figshare.4029480.v129
Figshare: Sonification of phantom image volume. doi: 10.6084/m9.figshare.4029495.v130
Figshare: Sonification of focalcheck beads. doi: 10.6084/m9.figshare.4029513.v131
Figshare: Sonification of Arabidopsis Plastid and Cell Membrane. doi: 10.6084/m9.figshare.4029519.v132
Figshare: Sonification of Arabidopsis Mitochondria and Cell Membrane. doi: 10.6084/m9.figshare.4029522.v133
Aprameya Mysore implemented and applied the method generated artificial data, and wrote the paper. Andreas Velten collected real data, guided the development of data generation and sonification methods, and helped writing the paper. Kevin Eliceiri initiated and oversaw the project.
The authors acknowledge technical support from the Laboratory for Optical and Computational Instrumentation (LOCI). The authors also thank Alexandra Chanoca and Marisa Otegui for providing the plants samples and Curtis Rueden for assistance with the FIJI implementation.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
References
1. Kollmeier B, Brand T, Meyer B: Perception of Speech and Sound. Springer Handbook of Speech Processing, Springer Science and Business Media. 2008; 65.Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 25 Oct 16 |
read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)