Keywords
Predictive Model, Multivariate Feature Selection, Hill Climbing, Random Forest
Predictive Model, Multivariate Feature Selection, Hill Climbing, Random Forest
Prostate cancer is the most common cancer affecting men. It is also one of the main causes of cancer mortality1. In addition to radiotherapy, androgen deprivation therapy (ADT) is a standard treatment. However, approximately one third of all patients with metastatic disease treated with ADT develop resistance to ADT. This condition is called metastatic castrate-resistant prostate cancer (mCRPC)1,2. Patients who do not respond to hormone therapy are often treated with a chemotherapy drug called docetaxel. The Prostate Cancer DREAM Challenge is a crowd-sourcing effort that aims to improve the prediction of survival and toxicity of docetaxel treatment in patients with mCRPC2. Specifically, there are two sub-challenges: 1) to predict overall survival of mCRPC patients using clinical trial data, and 2) to predict discontinuation of the docetaxel treatment due to adverse event at early time points. This paper reports our team’s effort contributing to sub-challenge 2.
The data for the challenge were provided by Project Data Sphere3, consisting of four clinical trials (ASCENT-24, VENICE5, MAINSAIL6, and ENTHUSE-337) for patients with mCRPC treated by docetaxel. The training data made available to the challenge participants consisted of 1600 patients from three clinical trials (ASCENT-2, VENICE, MAINSAIL). The clinical data from ENTHUSE-33 serve as the scoring set to test the prediction accuracy of submissions and hence, were not available to participants. Each team was allowed a maximum of two online submissions. In our two submissions, we used the same methods but varied the hold-out data8,9. After the challenge window, the submission portal was re-opened, allowing participants to continue their effort in refining and exploring alternative methods.
As a part of the Prostate Cancer DREAM Challenge, we developed data-driven models to predict patient outcomes in mCRPC with subsequent discontinuation of docetaxel therapy. We contributed to sub-challenge 2, which aims to predict discontinuation of docetaxel treatment due to adverse events. We empirically studied and assessed the performance of various machine learning algorithms and feature selection methods using cross validation on the provided training data. We assessed our predictive models using the area under the curve (AUC)10, which is the scoring metric adopted by the Prostate Cancer DREAM Challenge sub-challenge 2. This paper reports the predictive models we developed for the Prostate Cancer DREAM Challenge as well as further improvements we made after the challenge was closed. The methods and our challenge submission are available online from Synapse8,9, and our post-challenge efforts are available on the GitHub repository11.
The training data consist of clinical variables across 1,600 mCRPC patients in three clinical trials, namely ASCENT-2 (Novacea, provided by Memorial Sloan Kettering Cancer Center, with 476 patients)4, VENICE (Sanofi, with 598 patients)5, and MAINSAIL (Celgene, with 526 patients)6. Specifically, longitudinal data from five tables were summarized into a core table consisting of 131 variables. The five tables of raw longitudinal data at patient-level include PriorMed (prior medication table records), Med-History (medical history table records patient reported diagnoses at time of patient screening), LesionMeasure (lesion table records target and non-target lesion measurement), LabValue (lab test table includes all lab data), and VitalSign (vital sign table records patient vital sign such as height and weight)2. We used the training data in the core table to build models predictive of treatment discontinuation (binary) for patients in a fourth clinical trial (test data), ENTHUSE-33 (AstraZeneca, with 470 patients)7.
Our approach consists of four main steps: (1) data cleansing and pre-processing, (2) feature selection, (3) classification, and (4) assessment, as shown in Figure 1.
Our analysis focused on the core data only. From the 1,600 patients, we removed 111 patients without clear discontinuation status, leaving a total of 1,489 patients across three clinical trials: ASCENT-2 with 476 patients, MAINSAIL with 420 patients, and VENICE with 593 patients. Data cleansing was performed separately within each clinical trial and later concatenated back together. Some features were only available on certain clinical trials, for instance, smoking frequency, which was only available on ASCENT-2. In contrast, features such as sodium, phosphorus, and albumin were available in two clinical trials other than ASCENT-2. The heterogeneity in the data also manifested in the different interpretation of the values. To name a few, features such as lesion locations HEAD_AND_NECK or STOMACH, only contained positive responses. In this case, we assigned all of the missing values as unknown, instead of negative values.
We kept variables that are available from all three clinical trials in the training data. We also performed data imputation using the impute package version 1.42.0 from Bioconductor12. Data imputation was applied to the missing data in the baseline lab values (such as alkaline phosphatase, etc.) using the k nearest neighbor (KNN) algorithm. We varied the value of k from 1 to the number of patients, and evaluated the performance against a naive-bayes classifier13 in a 10 cross-fold validation. k = 40 was shown to be an optimal parameter. Missing information such as the patients’ weight, height and BMI were replaced by the average value of all patients. For the discrete variables, we replaced the missing values with a new variable (‘unknown’ value). Data augmentation was also performed by converting selected multi-label variables into binary variables, such as ‘smoking frequency’. Figure 2 shows how the split and reconstruction was performed.
To avoid over-fitting, we performed data cleansing and data pre-processing for the testing data (ENTHUSE-33) separately.
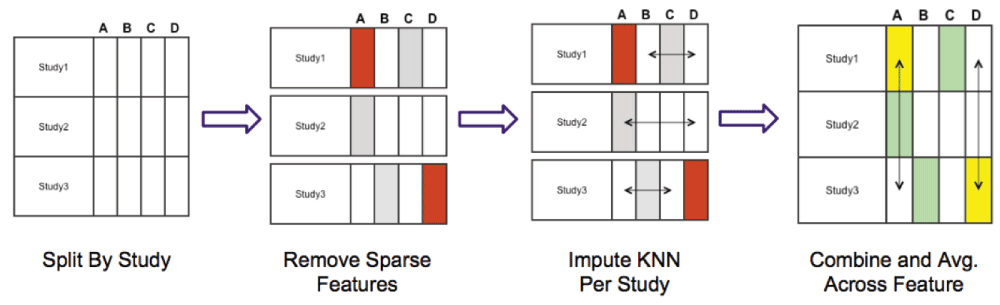
In step 2 (Remove Sparse Features) considering each dataset in turn, we removed features that were missing ample data (>79%) to prevent KNN imputation (illustrated as red blocks). In step 3, also considering each clinical trial as distinct, we used KNN to impute missing data from the features that contained missing data (illustrated as grey blocks). Finally, in step 4 all datasets were concatenated. and the features removed in step 2 (illustrated now as yellow blocks) were replaced by the mean values of that feature calculated from the other two clinical trials (shown using vertical arrows).
We observed that univariate feature selection methods did not perform well in this case. We used cross validation to guide us in the search for relevant features (or clinical variables) in the data. Specifically, we assessed our models using the area under the precision-recall curve (AUC) using the ROCR R package version 1.0-710.
We adopted the multivariate hill-climbing14 approach that optimized the AUC using 10-fold cross validation of the training data to search for relevant features among 131 features (clinical variables). The algorithm started with a random set of features against the model and returned the AUC. Depending on the AUC, the algorithm removed, kept, or added other features, and iterated until it converged. This method was used in both of our submissions to sub-challenge 2. Hill-climbing aims to maximize accuracy and is a greedy approach. However, hill-climbing also has its limitations: it is computationally intensive and prone to be stuck at a local optimum. In addition, it also tends to converge with different sets of features within each cross validation run, which makes it difficult to determine what are the corresponding factors that contribute to the discontinuation of the treatment. As one of the goals of the challenge is to identify prognostic markers in patients with mCRPC who will discontinue the treatment, hill-climbing may not the ideal approach.
Halabi et al. reported a list of strong predictors for overall survival of mCRPC patients15,16. These predictors include race, age, BMI, prior radio therapy, prior analgesics, and patient performance status. Lab results for albumin, lactate dehydrogenase, white blood cells, aspartate amminotransferase, total bilirubin, platelet count, hemoglobin, alanine transaminase, prostate specific antigen and alkaline phosphatase were also reported to be strong predictors of overall survival for patients with mCRPC. We hypothesize that the underlying molecular mechanisms that drive overall survival (the goal of sub-challenge 1) and treatment discontinuation (the goal of sub-challenge 2) are related. In addition, after the challenge was closed and the winners were announced, we checked out the winners’ winning strategy. In particular, we were inspired by Team Jayhawks from University of Kansas Medical Center who stated that they "made use of the variables we derived for sub-challenge 1a and also the overall risk score for survival". Therefore, we experimented with the set of features for overall survival reported by Halabi et al. and used the results as the baseline. Additionally, we also performed random forest importance from FSelector package17 to look for additional features to improve the prediction accuracy for treatment discontinuation (sub-challenge 2).
Random forest measures the importance of a variable by estimating the prediction error when that variable is permuted and the rest of variables remain unchanged. We evaluated the importance of the remaining variables using random forest. By varying the number of the features and evaluating the results, medical history (neoplasms benign, malignant or unspecified), smoke, and glucose were identified as the contributing factors. In modeling this feature selection, we addressed the issue of imbalanced data by performing random sampling using the training data consisting of 0.31 positive samples (patients known to have discontinued the treatments) and 0.69 negative samples.
We applied various classification algorithms to the selected features, including support vector machine (SVM)18, decision trees19, neural networks20, random forest21,22, and ensemble methods23. We observed comparable performance across different classification methods, and subsequently selected random forest as the classifier in our final submissions due to its robustness in heterogeneous datasets. As an ensemble method, random forest splits the training data into a number of subset and constructed decision trees as the classification model. Although the random forest package from CRAN21 usually comes with a set of sensible default parameters, we performed cross validation to optimize the classifier models. Subsequently, we adopted the following set of parameters: pick 9 random variables on each split (parameter mtry), and set the number of trees to 6,300 trees (parameter ntree). During all of the model tuning, 123 was used as the random seed.
For sub-challenge 2, each team was allowed 2 submissions. In our two submissions, we used the same methods and varied the hold-out data8,9. See Table 1 for detailed results.
Each column represents a different feature selection technique and/or hold-out dataset. Columns labeled ‘ENTHUSE-33’ were the test set provided by the Challenge organizers, and hence, were scored via the DREAM 9.5 submission system, while others were scored using the hold-out study as the testing dataset. BL = baseline (features selected based on the Halabi paper only16, HC = hill-climbing, RFI = combining features identified by Halabi and random forest importance.
We applied hill-climbing to select features from the model space and random forest as the classification methods by varying the hold-out data (see Table 1). We performed 10-fold cross validation by randomly selecting 10% of the training data across all three clinical trials as the hold-out data, and repeated the procedures 10 times. This 10-fold cross validation procedure using hill-climbing yielded an average AUC of 0.532. However, we achieved a substantial reduction in AUC (0.129) when we applied this model to the scoring data (470 patients from ENTHUSE-33 clinical trials). This model served as our 1st submission and ranked 35th on the leaderboard.
We went on to conduct additional empirical experiments to identify the difference between AUC from 10-fold cross validation and AUC from the scoring set. We hypothesized that this difference in AUCs from cross validation and from the scoring data is due to heterogeneity in the data collected in different clinical trials. Therefore, we studied the heterogeneity of three clinical trials in the training data by using each of three clinical trials as hold-out data. Table 1 showed that using the VENICE clinical trials as the hold-out data resulted in AUCs that are comparable to what we observed in our 1st submission. In particular, we produced an AUC of 0.171 by holding-out the VENICE clinical trial in the training data. Our 2nd submission resulted from applying hill-climbing and random forest to the ASCENT-2 and MAINSAIL clinical trials, and achieved an AUC of 0.132 on the scoring data (ENTHUSE-33). Our 2nd submission ranked 34th out of 61 submissions on the leaderboard. The AUC measured by the top performer was 0.19024.
Figure 4 shows that albumin was consistently selected by hill-climbing as a strong predictor disregard to the hold-out data. In total, there are 27 clinical variables selected in more than one hill-climbing model, including: Na (sodium), OTHER (other lesion), ALB (albumin), ORCHIDECTOMY, CEREBACC cerebrovascular accident either hemorrhagic and or ischemic), AST (aspartate aminotransferase), HB (hemoglobin), Mg (magnesium), LYMPH_NODES (lesion), PROSTATE (lesion), BILATERAL_ORCHIDECTOMY, CORTICOSTEROID (medication), CREAT (creatinine), PSA (prostate specific antigen), CREACL (creatinine clearance).
After the challenge ended, we continued to fine tune our models and submitted predictions to be scored. We adopted the Halabi model16 and combined it with random forest importance (RFI) to improve prediction. Random forest was kept as the classification method, and we also varied the hold-out data as the assessment. Random forest importance was computed by excluding features from the Halabi model which resulted in different sets of features depending on the hold-out data (see Figure 5).
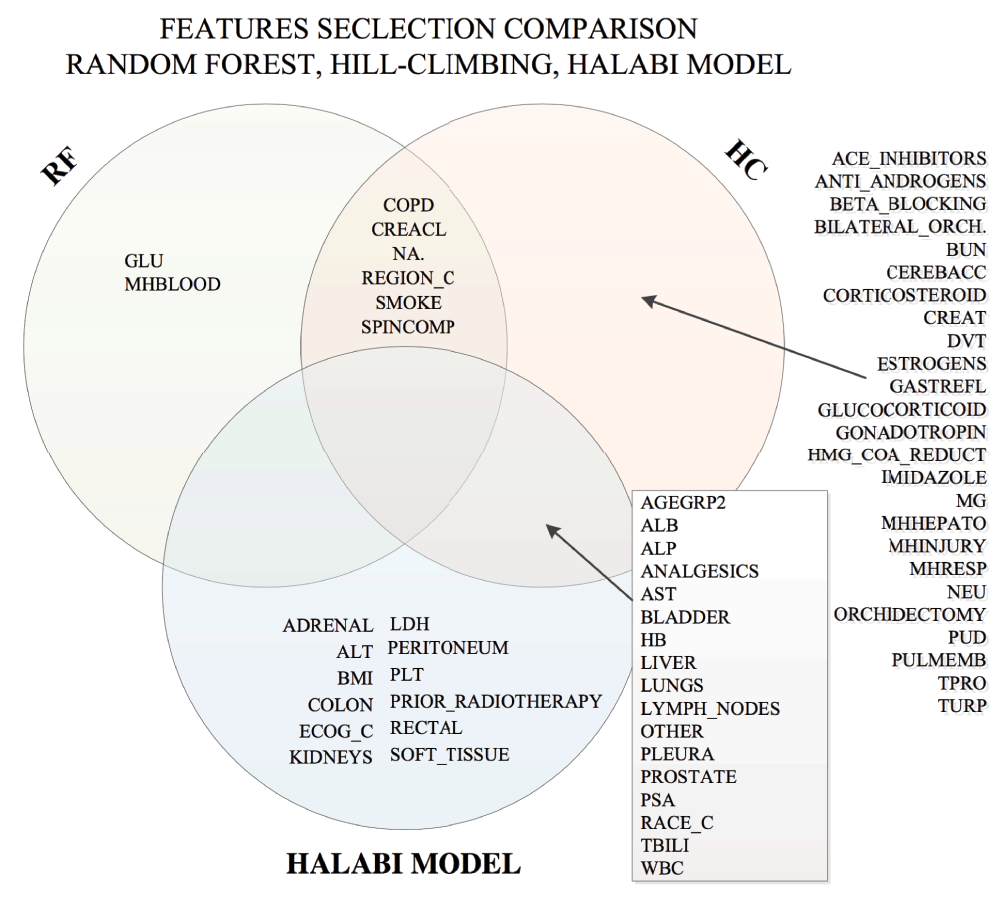
Next, we compared the features chosen by hill-climbing, RFI and the Halabi model. We observed that SMOKE and REGION overlapped among the hill-climbing and RFI results as shown in Figure 6. By varying the number of top features ranked by random forest importance combined with the Halabi model, 4 top ranking clinical variables resulted from random forest importance yielded the best average accuracy (see Figure 3). Applying this predictive model in 10-fold cross validation resulted in an average AUC of 0.275. We also repeated the process by selecting each individual clinical trial as the hold-out data, which yielded AUCs of 0.106 (VENICE as the hold-out), 0.303 (ACCENT-2), and 0.263 (MAINSAIL). Compared to the hill-climbing method, this model produced better and more consistent AUCs across clinical trials. Using each of the trained models to predict the discontinuation of docetaxel treatments for 470 patients from ENTHUSE-33 (AstraZeneca) clinical trials resulted in AUCs of 0.140 (VENICE), 0.132 (ACCENT-2), 0.124 (MAINSAIL), as also shown in Table 1.
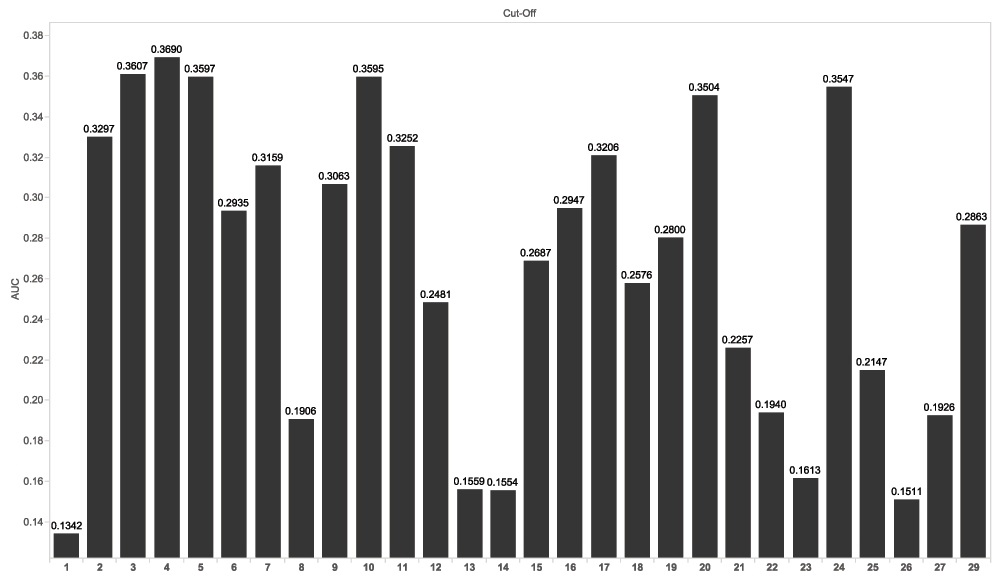
Random forest was used as the classification method, and 10% of randomly selected data across all sample as the holdout data.
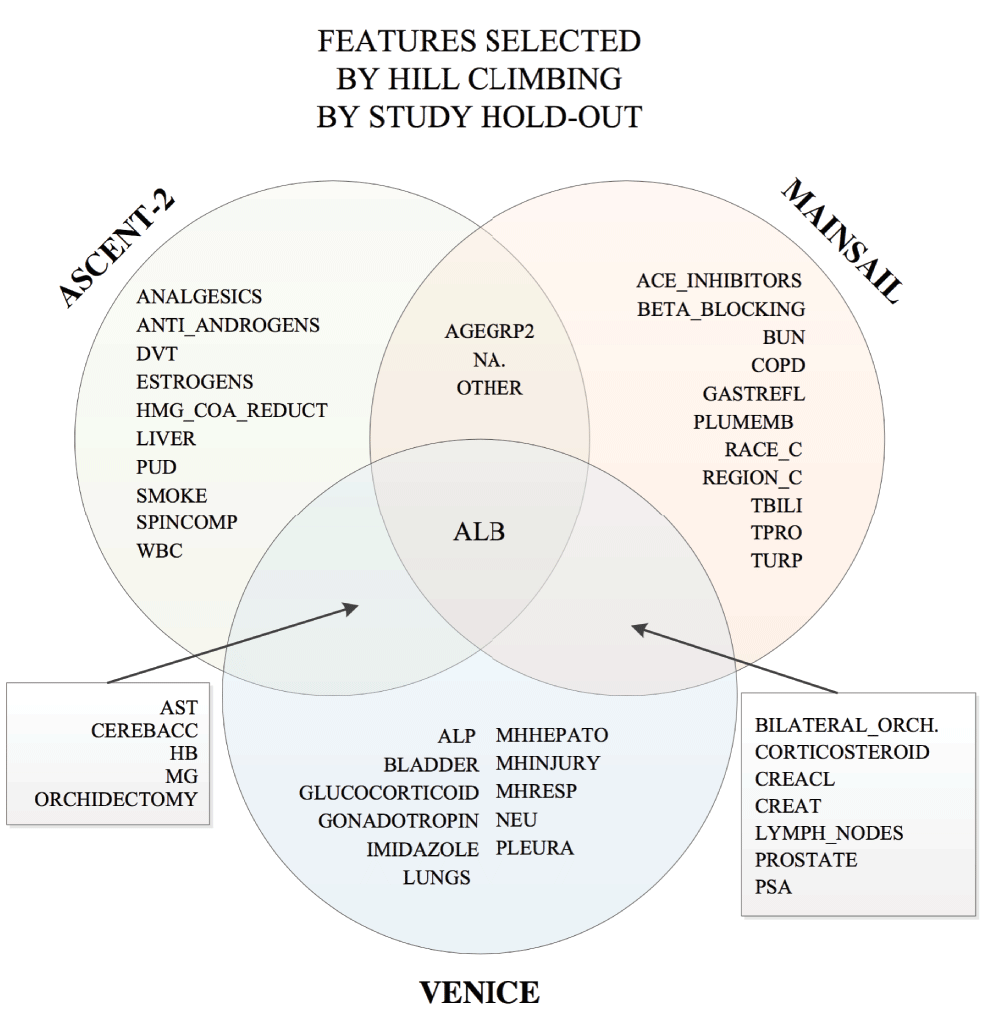
Random forest was used as the classification model.
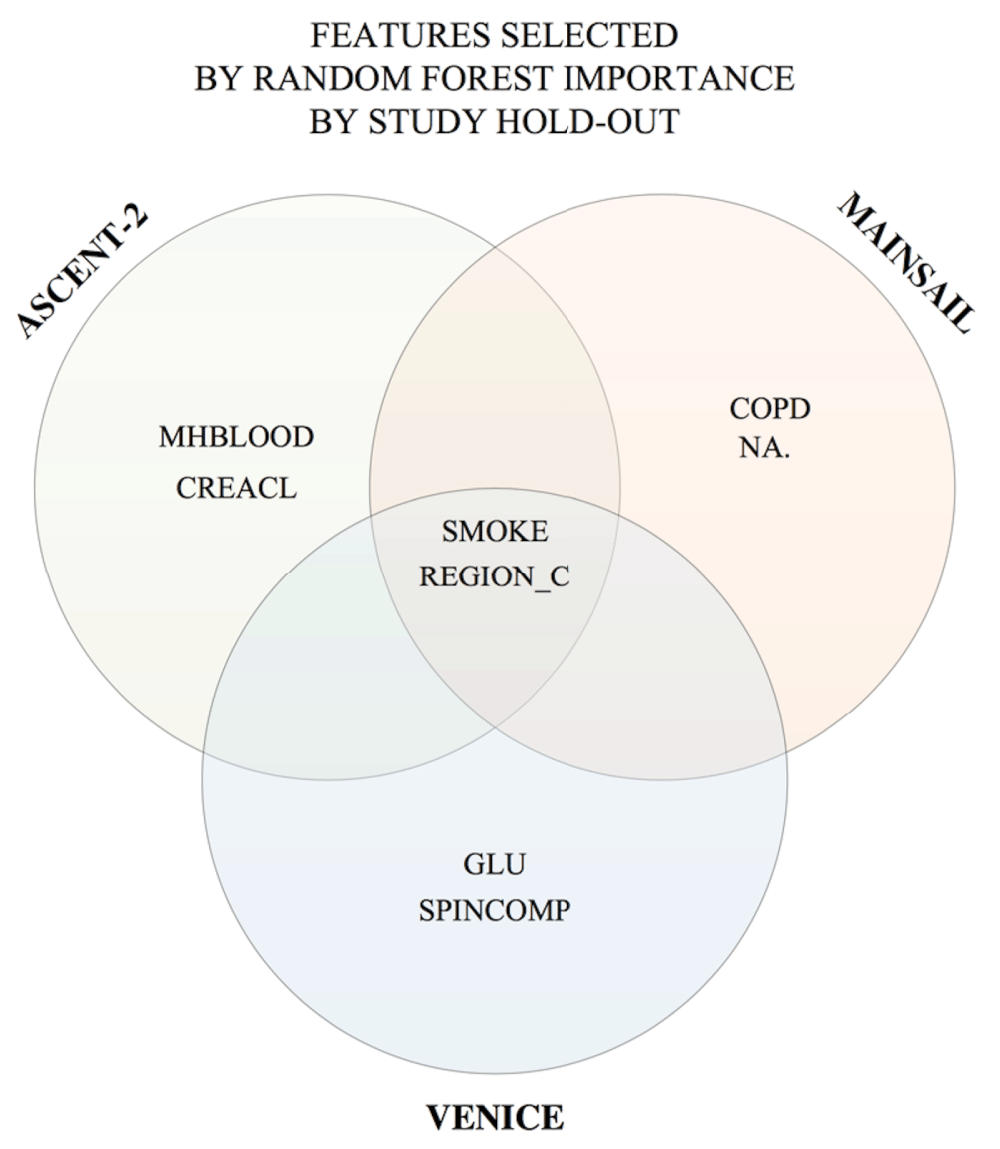
Features identified by the Halabi model was excluded during the computation.
Random forest was used as the classification method. RFI: the union of all features selected by random forest importance by varying the hold-out data after excluding features described by the Halabi model. HC: is the union of all features selected by hill-climbing by varying the hold-out data.
A major challenge of the Prostate Cancer DREAM Challenge was the unbalanced class sizes and the heterogeneity of the clinical trials. Subsequent to data cleansing, there remained only 197 positive samples (patients who discontinued the treatments) and 1,292 of negative samples. We observed that data cleaning and augmentation improved the AUC and F1 (before we augmented the data, our AUCs were in the range of 0.2 in 10-fold cross validation). Lastly, we were delighted to find that our computationally intensive hill-climbing algorithm, designed to find an optional feature set, provided strong results when testing using the datasets provided to us. When scored using the ENTHUSE-33 dataset, hill-climbing performed weakly against the combination of Halabi model and random forest importance.
We experimented with various feature selection and classification methods. We observed that some clinical variables were selected more consistently across feature selection methods, see Figure 6. These variables include: AGEGRP2 (Age Group), ALB (Albumin), ALP (Alkaline Phosphatase), ANALGESICS, AST (Aspartate Aminotransferase), BLADDER (Lesion), COPD (Blood and Lymphatic System), CREACL (Creatinine Clearance), HB (Hemoglobin), LIVER (Leison), LUNGS (Lesion), LYMPH_NODES (Lesion), NA. (Sodium), OTHER (Other Lesion), PLEURA (Pleura), PROSTATE (Lesion), PSA (Prostate Specific Antigen), RACE_C, REGION_C, SMOKE, SPINCOMP (Spinal Cord Compression), TBILI (Total Bilirubin), and WBC (White Blood Cells).
In this study, we only looked at the core table that was precompiled from detailed longitudinal tables. Given more time and resources, a close look at the raw longitudinal data may yield additional insight in discovering the clinical variables that predict the discontinuation of treatment for mCRPC patients.
The Challenge datasets can be accessed at: https://www.projectdatasphere.org/projectdatasphere/html/pcdc Challenge documentation, including the detailed description of the Challenge design, overall results, scoring scripts, and the clinical trials data dictionary can be found at: https://www.synapse.org/ProstateCancerChallenge
The code and documentation underlying the method presented in this paper can be found at: http://dx.doi.org/10.7303/syn46018488 and http://dx.doi.org/10.7303/syn47297619. The method and results are also presented as a poster25.
DK served as team captain of team ‘yoda’ in the DREAM9.5 Prostate Challenge, wrote the first draft of the manuscript and was responsible for code consolidation and submission. KA contributed data cleansing scripts, created figures and assisted in the writing of this paper. KYY supervised the project through its duration. All authors contributed to the writing of the manuscript.
Ling Hong Hung and Ka Yee Yeung are supported by NIH grant U54-HL127624. Daniel Kristiyanto is sponsored by the U.S. Department of State and American-Indonesian Exchange Foundation (AMINEF) through Fulbright Scholarship, and gratefully acknowledges funding provided by the University of Washington in the form of full tuition waivers.
This publication is based on research using information obtained from www.projectdatasphere.org, which is maintained by Project Data Sphere, LLC. Neither Project Data Sphere, LLC nor the owner(s) of any information from the web site have contributed to, approved or are in any way responsible for the contents of this publication.
This project used computing resources provided by Microsoft Azure.
We also would like to thank all students in TCSS 588 Bioinformatics in the Spring Quarter of 2015 at University of Washington Tacoma who contributed to this project, and extend our acknowledgments to Sage Bionetworks, the DREAM organization, and Project Data Sphere for developing and supplying data for the challenge.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 16 Nov 16 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)