Keywords
3D printing, neurodegenerative disease, cell culture, in vitro release, mucopolysaccharidosis, corpus callosum
3D printing, neurodegenerative disease, cell culture, in vitro release, mucopolysaccharidosis, corpus callosum
Methods were updated to include alternative data file types that may be used.
To read any peer review reports and author responses for this article, follow the "read" links in the Open Peer Review table.
Mucopolysaccharidosis (MPS) is a spectrum of inheritable conditions involving the accumulation of glycosaminoglycans (GAGs) following disruption of key lysosomal enzymes, which in turn leads to complications on a cellular, tissue, and organ level1. In MPS type I (MPS I), which is characterized by a deficiency of the enzyme α-L-iduronidase, brain MRI scans reveal thinning of white matter and lesions within the periventricular area and especially the corpus callosum (CC)2. The CC is the largest white matter structure in the brain, with more than 300 million axonal projections, and it interconnects the left and right hemispheres3. MPS I leads to patient-specific, irregular white matter density and geometry in the CC. Current treatment for MPS include enzyme replacement therapy (ERT) and hematopoietic stem cell transplantation (HSCT). ERT has been shown to ameliorate MPS symptoms, yet does not prevent disease progression4, owing in part to poor bioavailability. HSCT has been shown to improve cognitive development. Donors, however, can be hard to find unless umbilical cord blood is available; the procedure also has significant health risks5,6. As such, more research into potential targets and drug delivery excipients, which can provide tunable release kinetics, is needed to develop a library of promising treatment options. Additionally, owing to the highly patient-specific deterioration of cerebral white matter, patient-specific identification of synergistic drug combinations and optimal drug release kinetics can enable a more personalized medicine approach to treat MPS in the future.
Traditional methods of screening use two-dimensional (2D) cell culture to study biochemical pathways and targets in cells. Yet, 2D designs of traditional cell cultures fail to account for complex cell-cell and/or cell-matrix interactions. There has been a growing literature that demonstrate the importance of three-dimensional (3D) environments in expressing phenotypes, genes, and proteins at levels found in vivo and not otherwise seen in 2D models7–9. 2D in vitro drug release studies of promising therapeutic targets are generally limited to providing qualitative insight into in vivo release behavior. Seemingly arbitrary choices in probing conditions such as material volume, material surface area, supernatant volume, and rotator conditions, hinders quantitatively rigorous conclusions of mass transfer, pharmacokinetic, and pharmacodynamics properties to be made from benchtop measurements. These in tandem demonstrate a pressing need for the use of 3D disease models as a more representative in vitro system. Here, we describe an inexpensive and fast method of developing such patient specific 3D models.
The 3D brain MRI scans of a 20-year-old male subject with MPS I and an age-matched healthy male control were manually traced to obtain a 3D structure of the corpus callosum (CC). The 3D model was printed on a Makerbot Replicator 5X, sterilized (Figure 1), and could be used for cell culture or in vitro release studies. The de-identified MRI scans were obtained as Digital Imaging and Communications in Medicine (DICOM) files (Dataset 111). The CC was traced on the mid-sagittal slice and five adjacent slices in each hemisphere using open source InVesalius 3 (http://www.cti.gov.br/invesalius/, RRID: SCR_014693). Alternatively, OsiriX 8.0.1 software (http://www.osirix-viewer.com/, RRID: SCR_013618) may also be used. In some cases, data is obtained as a NIfTI-1file with the extension .nii; these files may also be used. MRICron 1.0 (http://people.cas.sc.edu/rorden/mricron/index.html) RRID:SCR_002403 or mri_convert 1.0 (https://surfer.nmr.mgh.harvard.edu/fswiki/mri_convert) software packages may be used to convert between DICOM and NIfTI-1. The software was then used to render the scans into a single .STL file (Dataset 212). The 3D model of the CC was loaded into MakerBot Desktop v. 3.6.0.78 (https://www.makerbot.com/download-desktop/) and printed on a MakerBot Replicator 5X with poly(lactic acid) at a resolution of 0.2 mm, maintaining life-size dimensions. Stratasys post-processing fluid was optionally used to remove any support material. The 3D printed structures were rinsed with a 70% ethanol/water solution and UV-sterilized overnight. The prints were then coated with polylysine (Sigma) for cellular adhesion10, by dipping them upside down in a 0.5 mg/mL poly-L-lysine solution for at least 10 minutes. Only the top of the surface was dipped (Figure 1d), as this was the area of interest where the drug delivery materials would be loaded, but for other applications discussed in the next section, the entire structure can be dipped into ~50 mL of the poly-L-lysine solution for complete cell adhesion on the top and bottom.
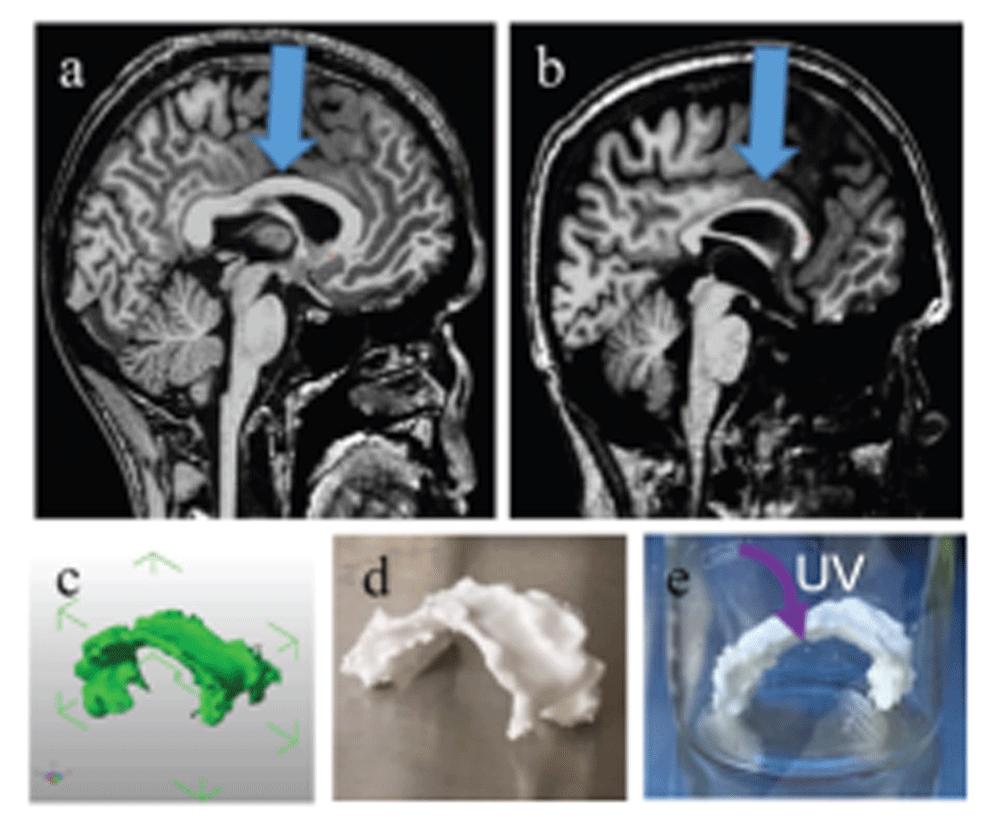
(a–b) T1-weighted brain MRI with resolution of 1×1×1 mm, midsagittal slice, arrow pointing at corpus callous in (a) healthy control and (b) MPS I subject. (c) CAD image of MPS I corpus callosum taken at five adjacent slices in each hemisphere. (d) 3D printed MPS I corpus callosum with poly(lactic-acid) on a Makerbot Replicator 5X. (e) UV sterilizing the print overnight.
For cell culture, this object would be used in a sterile flask, or alternatively, a larger sterilized container. For use as a drug delivery platform, the object could be both be cultured with cells as previously discussed and kept in cell culture media, or used without cells in PBS. A parenteral drug delivery system, such as shear-thinning hydrogels or hydrophobic polymer melts, can be injected with a syringe on top of the 3D model, until the amount of drug loaded is comparable to translational doses, or until the material completely coats the 3D model. When a cell coated surface is used, the drug will be released into the cell media. Conversely, when a cell-free model is used, the drug release kinetics are monitored in PBS, which is less prone to interference. Care must be taken to ensure that the release kinetic probing molecule’s absorbance or emission spectra are not greatly interfered with by cell culture media.
This technique uses, but is not limited to, poly(lactic-acid) (PLA), a readily available filament for desktop 3D printers, such as the Makerbot Replicator 5X. PLA has been widely shown to be biocompatible13. Applications of this platform include studying in vitro drug release of injectable drug depots for delivery of therapeutics e.g. proteins for enzymatic deficiency disorders, such as MPS, and hydrophobic small molecules for brain cancer. In vitro drug release methodology can largely vary release profiles depending on the geometry of the container used. A 15 mL conical tube provides a different area for mass transfer than a 10 cm culture dish or 5 mL glass vial. This 3D modeling platform can potentially offer a more realistic and more standard geometry for monitoring drug release. Additionally, many therapeutic approaches to treat brain cancer and other diseases rely on injecting or implanting material that maintains a high interfacial concentration to improve drug bioavailability and efficacy, such as gold nanoparticle radiosensitizers for radiotherapy14,15. The drug depot material can be tested in vitro on this platform to determine the proper interfacial concentration given to-scale surface area of the tissue, and monitor the duration to which this concentration can be maintained.
Figure 1 (a–b) demonstrates the thinning of the CC in MPS I (b) compared to that of a healthy brain (a). Given the uniqueness of each MPS patient’s brain pathology, density, and geometry, the ability to test the therapeutic window, effectiveness, and optimal drug loading concentration into an injectable drug depot for each specific patient is highly useful. A team of high school and undergraduate students was able to render the CAD file (Figure 1c) and 3D print on common desktop 3D printers at a low cost (Figure 1d), suggesting that this culture may be scaled more readily than expensive 3D in vitro platforms. This material’s modulus is approximately 3 GPa, several orders of magnitude larger than native tissue. In order to create a 3D cell culture platform which enables cell migration and proliferation within the tissue, a 3D bioprinting approach must be used16,17. In conclusion, this method’s robustness, ease, and low cost make it adaptable for use for a wide variety of applications in drug delivery, drug discovery, tissue engineering, and stem cell biology.
Dataset 1: DICOM files for the de-identified MRI scans of the corpus callosum of a MPS I subject, doi: 10.5256/f1000research.9861.d14432711
Dataset 2: Resulting CAD file from InVesalius 3 software (.STL), used to render the DICOM files in Dataset 1, doi: 10.5256/f1000research.9861.d14432812
The study protocol involving the brain MRI acquisition was approved by the University of Minnesota IRB committee. Written, informed consent to publish results from MPS patients and healthy volunteers was obtained.
AT, SS, and IN developed the experimental outline. AT, MG, SS, SC, AM, DG, BH, and RT developed the model. AT, SS, AM, DG, and IN wrote the first draft of the paper. All authors revised the manuscript and have agreed to the final content.
The MRIs were provided from the projected funded by Lysosomal Disease Network (RDCRN; grant number NIH U54NS065768). Parts of this work were funded by a CoCreate Community Research Grant (#16354).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors thank Nicholas Powley, Mac Cameron, and Heather Fong for encouragement in pursuing this project.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 16 Mar 17 |
||
|
Version 1 02 Dec 16 |
read | read |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)