Keywords
knockdown, siRNA, negative control, specificity, Western Blot, shRNA, transfection, vector
This article is included in the Antibody Validations gateway.
knockdown, siRNA, negative control, specificity, Western Blot, shRNA, transfection, vector
Poor quality, unverified antibodies are a major factor underlying the crisis of reproducibility in research1, resulting in an estimated $800 million of waste each year2 and a ten-fold spike in retractions over the past decade3. Behind those numbers are researchers with mounting frustration each time a new batch of reagents fails to reproduce their previous work. If we as a scientific community are to eliminate this problem plaguing our research, we must develop new and more thorough validation methods to fortify and standardize routine studies.
We invite industry to respond to the need for more rigorous validation by incorporating negative controls, an essential part of experimental design that has nonetheless been overlooked historically by antibody manufacturers and distributors. Companies typically validate with Western blots but seldom test routinely whether antibodies still produce a signal when the target protein is suppressed or removed.
Short interfering RNA (siRNA) knockdown is a technology that makes routine use of this strategy more feasible. This technology degrades target messenger RNA to ‘knock down’ the production of a protein in the cell. The combination of siRNA-treated cells and a specific antibody will result in a significant drop in signal compared to an untreated sample by Western blot (Figure 1).
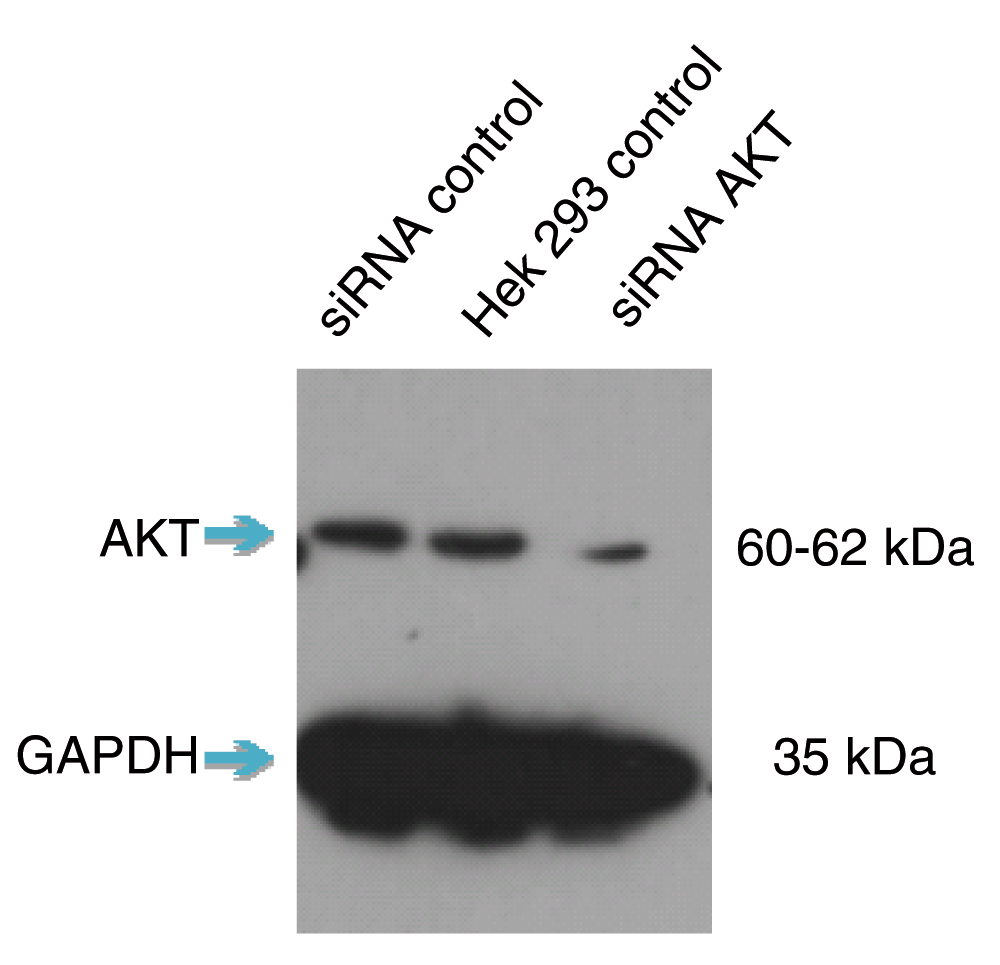
siRNA knockdown has not been the only validation initiative put forward to identify non-specific antibodies4. CRISPR and other gene editing methods knock out a gene from DNA, preventing the associated protein from ever being produced. By default, an antibody binding to any protein in this environment is binding to the wrong protein. Knock-out is a powerful negative control, however one concern with this method is the risk of cell death when a target protein is vital to cell survival. Other initiatives dispense with negative controls and verify positive identification instead. Mass spectrometry, for example, yields unique spectra that differentiate an antibody-bound protein of interest from any other bound molecule but is limited only to those proteins that can be immunoprecipitated.
While all of these approaches sound simple, their methodologies can be complex. Anyone seeking a deeper understanding of siRNA knockdown validation studies, or considering their own validation protocol, may find the following overview useful. These steps are based on Proteintech methods, which have been tested and refined over years. However, they are by no means the only way to perform this validation.
Selecting a target and designing an appropriate vector to transfect into cells is a relatively straightforward process, with an abundance of literature and online resources available to guide the process (RNAi Consortium, Dharmacon, Ui-Tei, and Genelink). The actual engineering, of course, requires a bit more hands-on finesse to create the short hairpin RNA that is the precursor form of the siRNA. The ultimate aim is to design two single-stranded 19-22mer DNA oligonucleotides (one sense strand, one antisense strand) whose transcription products will eventually anneal together, linked at one end by a short loop sequence. Proteintech uses the loop sequence TTCAAGACG.
Once the vector has been generated and produced in sufficient quantity, scientists need to determine an appropriate transfection method for delivery into cells. Upon successful transfection, the cells transcribe the foreign DNA to generate the shRNA described above (Figure 2). Afterward, Dicer processes the shRNA into siRNA by removing the loop sequence. The resulting siRNA binds with RISC (RNA-induced silencing complex), which separates the two strands of the RNA and activates the complex. RISC remains bound to one strand, that complementarily binds to a target mRNA and cleaves it. This suppresses production of the associated protein. Scientists need to allow time for the biological processes to be carried out, while culturing the cells to produce enough sample for a Western blot.
Cell death can be a major source of frustration, particularly if the target protein is vital to cell survival. Fortunately, most cells can propagate normally if the protein is suppressed, but not completely eliminated. In such cases, knockdown methods can be fine tuned and are likely a preferred alternative to knock out validation, which prevents any transcription through gene editing.
A strong signal for the empty-vector cells next to a weak signal for the siRNA-transfected cells in a Western blot means that the antibody is specific and that the knockdown experiment was successful. Any nonspecific bands should raise questions, as these could indicate that the antibodies themselves are nonspecific. Bands should also be consistent across all lanes of the Western blot membrane. If not, something may have gone wrong in the experiment, and protocol and experimental design should be reviewed.
While the value of negative controls to antibody validation protocols should not be understated, it is also understood that such validation is not widely available and that it consumes time and resources to perform in individual labs. Thorough Western blot validation with only positive controls can be a second-best alternative, provided that there is a sufficient quantity of data, and corroborated by published studies that reference the reagent catalog number and manufacturer. We encourage the development and use of negative controls such as siRNA knockdown as the new standard in antibody validation. Doing so will elevate reproducible research and accelerate scientific progress.
WO wrote and JL critically reviewed and edited the article. All authors have agreed to the final content of the manuscript.
Will Olds is a scientific officer at Proteintech Group. Jason Li is CEO of Proteintech Group.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 09 Mar 16 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
The method of publishing information about an antibody. It does not matter how much you validate if the ... Continue reading This is a well written piece, but you have forgotten one important part.
The method of publishing information about an antibody. It does not matter how much you validate if the antibody you are validating is not identified sufficiently to actually determine which antibody is being used!
For your 10176-2-AP antibody, that you use as an example. Please add the RRID to it so that the data is kept "safe" and uses of this reagent are tracked through the literature.
Please see the recommendations from F1000 instructions to authors: http://f1000.com/resources/F1000Research-Antibody-Validation-Article-Template.docx
- which states "Research Resource identifier (RRID) for each antibody, model organism, or software tool used, as can be found on http://scicrunch.com/resources"
The method of publishing information about an antibody. It does not matter how much you validate if the antibody you are validating is not identified sufficiently to actually determine which antibody is being used!
For your 10176-2-AP antibody, that you use as an example. Please add the RRID to it so that the data is kept "safe" and uses of this reagent are tracked through the literature.
Please see the recommendations from F1000 instructions to authors: http://f1000.com/resources/F1000Research-Antibody-Validation-Article-Template.docx
- which states "Research Resource identifier (RRID) for each antibody, model organism, or software tool used, as can be found on http://scicrunch.com/resources"