Keywords
Cancer Cell Lines, Pharmacogenomics, High-Throughput Screening, Biomarkers, Drug Response, Experimental Design, Bioinformatics, Statistics
This article is included in the Preclinical Reproducibility and Robustness gateway.
Cancer Cell Lines, Pharmacogenomics, High-Throughput Screening, Biomarkers, Drug Response, Experimental Design, Bioinformatics, Statistics
Pharmacogenomic studies correlate genomic profiles and sensitivity to drug exposure in a collection of samples to identify molecular predictors of drug response. The success of validation of such predictors depends on the level of noise both in the pharmacological and genomic data. The groundbreaking release of the Genomics of Drug Sensitivity in Cancer1 (GDSC) and Cancer Cell Line Encyclopedia2 (CCLE) datasets enables the assessment of pharmacogenomic data consistency, a necessary requirement for developing robust drug sensitivity predictors. Below we briefly describe the fundamental analytical differences between our initial comparative study3 and the recent assessment of pharmacogenomic agreement published by the GDSC and CCLE investigators4.
The first GDSC and CCLE studies were published in 2012 and the investigators of both studies have continued to generate data and to release them publicly. One would imagine that any comparative study would use the most current versions of the data. However, the authors of the reanalysis used an old release of the GDSC (July 2012) and CCLE (February 2012) pharmacological data, resulting in the use of outdated IC50 values, as well as missing approximately 400 new drug sensitivity measurements for the 15 drugs screened both in GDSC and CCLE. Assessing data that are three years old and which have been replaced by the very same authors with more recent data seems to be a substantial missed opportunity. It raises the question as to whether the current data would be considered to be in agreement and which data should be used for further analysis.
Given the complexity and high dimensionality of pharmacogenomic data, the development of drug sensitivity predictors is prone to overfitting and requires careful validation. In this context, one would expect the most significant predictors derived in GDSC to accurately predict drug response in CCLE and vice versa. This will be the case if both studies independently produce consistent measures of both genomic profiles and drug response for each cell line. In our comparative study3, we made direct comparison of the same measurements generated independently in both studies by taking into account the noise in both the genomic and pharmacological data (Figure 1a). By investigating the authors’ code and methods, we identified key shortcomings in their analysis protocol, which have contributed to the authors’ assertion of consistency between drug sensitivity predictors derived from GDSC and CCLE.
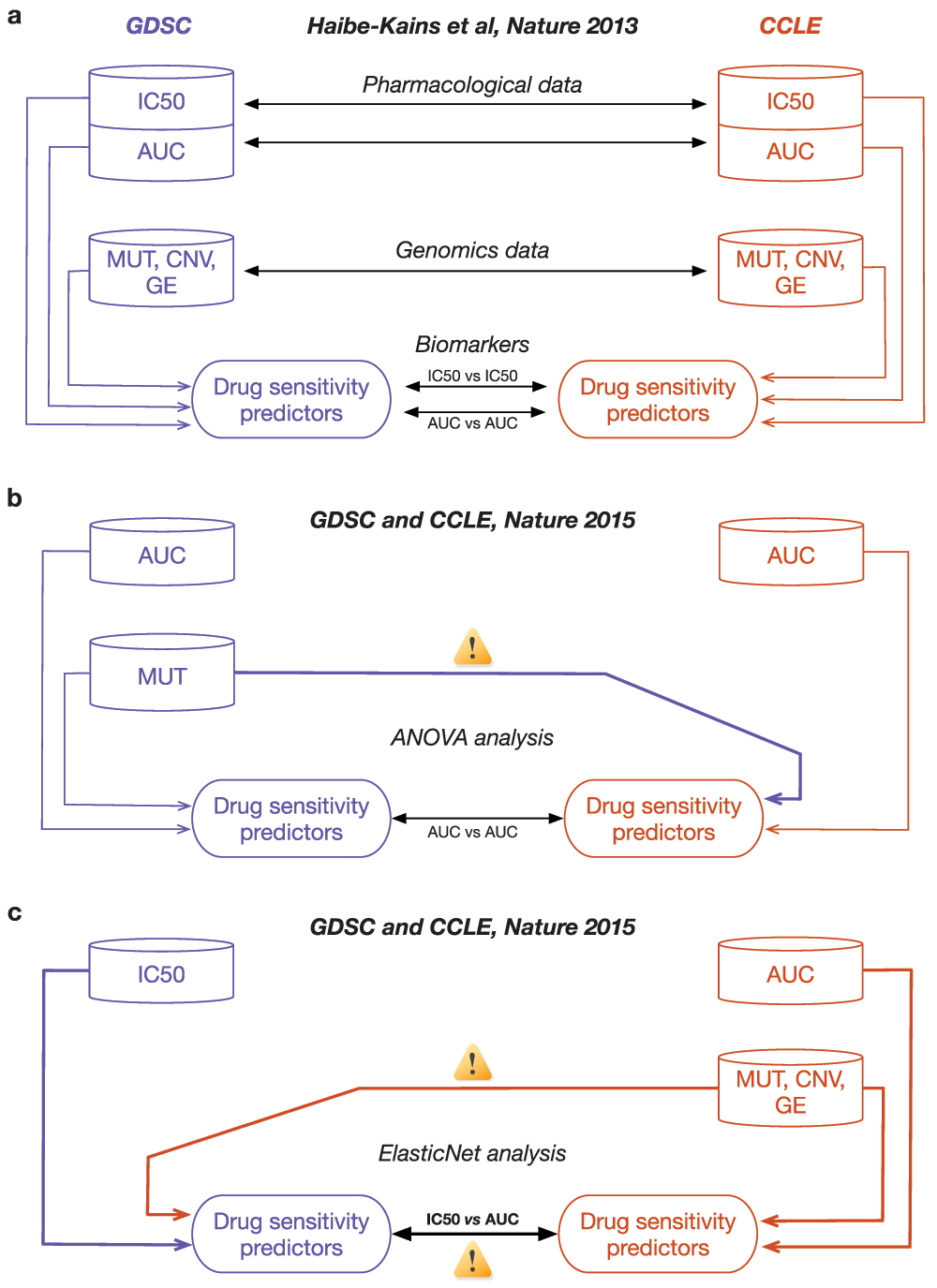
(a) Analysis design used in our comparative study (Haibe-kains et al., Nature 2013) where each data generated by GDSC and CCLE are independently compared to avoid information leak and biased assessment of consistency. (b) Analysis design used by the GDSC and CCLE investigators for their ANOVA analysis where the mutation data generated with GDSC were duplicated for use in the CCLE study. (c) Analysis design for the ElasticNet analysis where the molecular profiles from CCLE were duplicated in the GDSC study and the GDSC IC50 were compared to CCLE AUC data. Differences between our analysis design and those used by the GDSC and CCLE investigators are indicated by yellow signs with exclamation mark symbol.
For their ANOVA analyses, the authors used drug activity area (1-AUC) values independently generated in GDSC and CCLE, but used the same GDSC mutation data across the two different datasets (Figure 1b; see Methods). By using the same mutation calls for both GDSC and CCLE, the authors have disregarded the noise in the molecular profiles, while creating an information leak between the two studies. For their ElasticNet analysis, the authors followed a similar design by reusing the CCLE genomic data across the two datasets, but comparing different drug sensitivity measures that are IC50 in GDSC vs. AUC in CCLE (Figure 1c; see Methods).
We are puzzled by the seemingly arbitrary choices of analytical design made by the authors, which raises the question as to whether the use of different genomic data and drug sensitivity measures would yield the same level of agreement. Moreover, by ignoring the (inevitable) noise and biological variation in the genomic data, the authors’ analyses is likely to yield over-optimistic estimates of data consistency, as opposed to our more stringent analysis design3.
In examining correlation, there is no universally accepted standard for what constitutes agreement. However, the FDA/MAQC consortium guidelines define good correlation for inter-laboratory reproducibility5–8 to be ≥0.8. The authors of the present study used two measures of correlation, Pearson correlation (ρ) and Cohen’s kappa (κ) coefficients, but never clearly defined a priori thresholds for consistency, instead referring to ρ>0.5 as “reasonable consistency” in their discussion. Of the 15 drugs that were compared, their analysis found only two (13%) with ρ>0.6 for AUC and three (20%) above that threshold for IC50. This raises the question whether ρ~0.5–0.6 for one third of the compared drugs should be considered as “good agreement.” If one applies the FDA/MAQC criterion, only one drug (nilotinib) passes the threshold for consistency.
Similarly, the authors referred to the results of their new Waterfall analysis as reflective of “high consistency,” even though only 40% of drugs had a κ≥0.4, with five drugs yielding moderate agreement and only one drug (lapatinib) yielding substantial agreement according to the accepted standards9. Based on these results, the authors concluded that 67% of the evaluable compounds showed reasonable pharmacological agreement, which is misleading as only 8/15 (53%) and 6/15 (40%) drugs yielded ρ>0.5 for IC50 and AUC, respectively. Taking the union of consistency tests is bad practice; adding more sensitivity measures (even at random) would ultimately bring the union to 100% without providing objective evidence of actual data agreement.
The authors acknowledged that the consistency of pharmacological data is not perfect due to the methodological differences between protocols used by CCLE and GDSC, further stating that standardization will certainly improve correlation metrics. To test this important assertion, the authors could have analyzed the replicated experiments performed by the GDSC using identical protocols to screen camptothecin and AZD6482 against the same panel of cell lines at the Wellcome Trust Sanger Institute and the Massachusetts General Hospital.
Our re-analyses3,10 of drug sensitivity data from these drugs found a correlation between GDSC sites on par with the correlations observed between GDSC and CCLE (ρ=0.57 and 0.39 for camptothecin and AZD6482, respectively; Figure 2 a,b). These results suggest that intrinsic technical and biological noise of pharmacological assays is likely to play a major role in the lack of reproducibility observed in high-throughput pharmacogenomic studies, which cannot be attributed solely to the use of different experimental protocols.
In their comparative study, the authors did not assess the consistency of genomic data between GDSC and CCLE4. Consistency of gene copy number and expression data were significantly higher than for drug sensitivity data (one-sided Wilcoxon rank sum test p-value=3×10-5; Figure 3), while mutation data exhibited poor consistency as reported previously11. The very high consistency of copy number data is quite remarkable (Figure 3a) and could be partly attributed to the fact that CCLE investigators used their SNP array data to compare cell line fingerprints with those of the GDSC project prior to publication and removed the discordant cases from their dataset2.
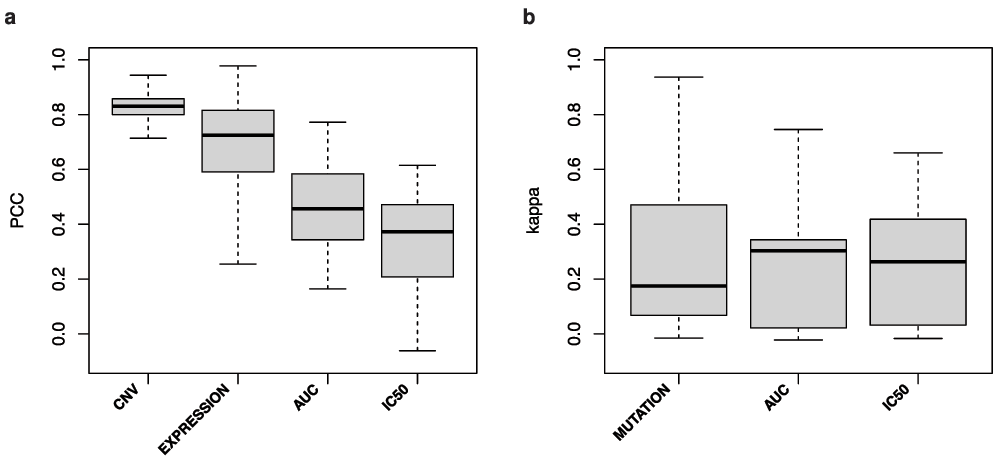
(a) Continuous values for gene copy number ratio (CNV), gene expression (EXPRESSION), AUC and IC50 and (b) for binary values for presence/absence of mutations (MUTATION) and insensitive/sensitive calls based on AUC >= 0.2 and IC50 > 1 microMolar values. PCC: Pearson correlation coefficient; Kappa: Cohen's Kappa coefficient.
We agree with the authors that their and our observations “[…] raise important questions for the field about how best to perform comparisons of large-scale data sets, evaluate the robustness of such studies, and interpret their analytical outputs.” We believe that a principled approach using objective measures of consistency and an appropriate analysis strategy for assessing the independent datasets is essential. An investigation of both the methods described in the manuscript and the software code used by the authors to perform their analysis4 identified fundamental differences in analysis design compared to our previous published study3. By taking into account variations in both the pharmacological and genomic data, our assessment of pharmacogenomic agreement is more stringent and closer to the translation of drug sensitivity predictors in preclinical and clinical settings, where zero-noise genomic information cannot be expected.
Our stringent re-analysis of the most updated data from the GDSC and CCLE confirms our 2013 finding that the measures of drug response reported by these two groups are not consistent and have not improved substantially as the groups have continued generating data since 201210. While the authors make arguments suggesting consistency, it is difficult to imagine using these post hoc methods to drive discovery or precision medicine applications.
The observed inconsistency between early microarray gene expression studies served as a rallying cry for the field, leading to an improvement and standardization of experimental and analytical protocols, resulting in the agreement we see between studies published today. We are looking forward to the establishment of new standards for large-scale pharmacogenomic studies to realize the full potential of these valuable data for precision medicine.
The authors’ software source code. As the authors’ source code, we refer to the ‘CCLE.GDSC.compare’ (version 1.0.4 from December 18, 2015) and DRANOVA (version 1.0 from October 21, 2014) R packages available from http://www.broadinstitute.org/ccle/Rpackage/.
As evidenced in the authors' code (lines 20 and 29 of CCLE.GDSC.compare::PreprocessData.R), they used GDSC and CCLE pharmacological data released on July 2012 and February 2012, respectively. However the GDSC released updated sets of pharmacological data (release 5) on June 2014, gene expression arrays (E-MTAB-3610) and SNP arrays (EGAD00001001039) on July 2015. CCLE released updated pharmacological data on February 2015, the mutation and SNP array on October 2012, and the gene expression data, on March 2013. These updates substantially increased the overlap in genomic features between the two studies, thus providing new opportunities to investigate the consistency between GDSC and CCLE10.
In the authors’ ANOVA analyses, identical mutation data were used for both GDSC and CCLE studies as can be seen in the authors’ analysis code in lines 20, 25–35 of CCLE.GDSC.compare::plotFig2A_biomarkers.R.
In their EN analyses, the authors compared different drug sensitivity measures, using IC50 in GDSC and AUC in CCLE, as described in the Supplementary Data 5 and stated in the Methods section of their published study:
“Since the IC50 is not reported in CCLE when it exceeds the tested range of 8 μM, we used the activity area for the regression as in the original CCLE publication. We also used the values considered to be the best in the original GDSC study: the interpolated log(IC50) values.”
This was confirmed by looking at the authors’ analysis code, lines 83 and 102 of CCLE.GDSC.compare::ENcode/prepData.R. Moreover, identical genomic data were used for both GDSC and CCLE studies, as described the Methods section of the published study:
This was confirmed by looking at the authors’ analysis code, lines 17, 38, 51, and 70 of CCLE.GDSC.compare::ENcode/genomic.data.R, and lines 10-11 of CCLE.GDSC.compare::plotFigS6_ENFeatureVsExpected.R.
All analyses were performed using the most updated version of the GDSC and CCLE pharmacogenomic data based on our PharmacoGx package12 (version 1.1.4).
All analyses were performed using the most updated version of the GDSC and CCLE pharmacogenomic data based on our PharmacoGx package12 (version 1.1.4). PharmacoGx provides intuitive function to download, intersect and compare large pharmacogenomics datasets. The PharmacoSet for the GDSC and CCLE datasets are available from pmgenomics.ca/bhklab/sites/default/files/downloads/ using the downloadPSet() function. The code and the data used to generate all the results and figures are available as Data Files 1 and 2. The code is also available on GitHub: github.com/bhklab/cdrug-rebuttal.
In the Methods, the authors use all cell lines to optimally identify the inflection point in the response distribution curves. The authors stated that “This is a major difference to the Haibe-Kains et al. analysis, as that analysis only considered the cell-lines in common between the studies when generating response distribution curves.” This is not correct. As can be seen in our publicly available R code, we performed the sensitivity calling (using the Waterfall approach as published in the CCLE study2 before restricting our analysis to the common cell lines, for the obvious reasons that the authors mentioned in their manuscript. See lines 308 and 424 in https://github.com/bhklab/cdrug/blob/master/CDRUG_format.R.
Open Science Framework: Dataset: Assessment of pharmacogenomic agreement, doi 10.17605/osf.io/47rfh13
Z Safikhani, N El-Hachem R Quevedo, and P Smirnov were responsible for downloading and curating the pharmacogenomic data. Z Safikhnao wrote most of the analysis code with the help of N El-Hachem R Quevedo, and P Smirnov. Z Safikhani, J Quackenbush and B Haibe-Kains designed the study. B Haibe-Kains supervised the study. All authors participated in the interpretation of the results. Z Safikhani, A Goldenberg, N Juul Birkbak, C Mason, C Hatzis, L Shi, H Aerts, J Quackenbush and B Haibe-Kains participated in the manuscript writing.
Z Safikhani was supported by the Cancer Research Society (Canada; grant #19271) and the Ontario Institute for Cancer Research through funding provided by the Government of Ontario. P Smirnov was supported by the Canadian Cancer Society Research Institute. C Hatzis was supported by Yale University. N Juul Birkbak was funded by The Villum Kann Rasmussen Foundation. C Mason was supported by the Starr Cancer Consortium grants (I7-A765, I9-A9-071), Irma T. Hirschl and Monique Weill-Caulier Charitable Trusts, Bert L and N Kuggie Vallee Foundation, WorldQuant Foundation (CEM), Pershing Square Sohn Cancer Research Alliance, NASA (NNX14AH50G), and the National Institutes of Health (R25EB020393, R01NS076465). L Shi was supported by the National High Technology Research and Development Program of China (2015AA020104), the National Natural Science Foundation of China (31471239), the 111 Project (B13016), and the National Supercomputer Center in Guangzhou, China. J Quackenbush was supported by grants from the NCI GAME-ON Cancer Post-GWAS initiative (5U19 CA148065) and the NHLBI (5R01HL111759). B Haibe-Kains was supported by the Gattuso Slaight Personalized Cancer Medicine Fund at Princess Margaret Cancer Centre.
The authors would like to thank the investigators of the Genomics of Drug Sensitivity in Cancer (GDSC) and the Cancer Cell Line Encyclopedia (CCLE) who have made their invaluable data available to the scientific community. We thank the MAQC/SEQC consortium for their constructive feedback.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 09 May 16 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)