Keywords
terpenoid metabolism, cannabis sativa, in silico gene expression, evolutionarily conserved genes,
This article is included in the Data: Use and Reuse collection.
terpenoid metabolism, cannabis sativa, in silico gene expression, evolutionarily conserved genes,
Due to its astonishing efficacy1, nowadays cannabis is prescribed by physicians for the treatment of neurological, psychiatric, immunological, cardiovascular, gastrointestinal, and oncological conditions2–7. Although therapies based on the use of cannabis flowers or their derivatives are recognized to be very effective, treatments centered on drugs containing Δ9-tetrahydrocannabinol (THC) alone lack efficacy and lead to increased side effects8,9. This discrepancy seems to result from the absence of the synergistic effects of additional pivotal compounds found in the Phyto-complex, the so-called entourage effect10. Among these molecules are other cannabinoids and terpenoids, which are thought to play major roles in the modulation of THC11.
Terpenes are small hydrocarbon (isoprenoid) molecules classified as either as monoterpenes, sesquiterpenes, diterpenes or carotenoids, depending on the number of isoprene units (C5) used to synthetize them. Terpenoids are small lipids derived from terpenes, often accompanied by a strong odor useful for the plants to protect themselves against possible predators12. Terpenoids not only have important functions when working in concert with cannabinoids; they have been widely investigated in many different plant species and are being exploited as anti-fungal, anti-bacterial, anti-oxidant, anti-inflammatory, anti-stress, anti-cancer and analgesic agents13–18. However, whilst the gene networks controlling the biosynthesis of cannabinoids and their precursors have been extensively studied19–22, the biosynthetic pathway of terpenoid molecules in Cannabis sativa is only recently being elucidated. Only two genes have been characterized, one encoding (-)-limonene synthase, the other (+)-α-pinene synthase23, two enzymes responsible for the conversion of geranyl pyrophosphate into limonene and pinene, respectively24,25. Remarkably, while cannabinoids are only found in cannabis plants, terpenoids are also produced by a variety of other plant species, so they must share many of the enzymes involved in their metabolism.
In the present work, evolutionarily conserved genes encoding enzymes predicted to be involved in terpenoid metabolism have been identified within the transcripts of the canSat3 reference transcriptome of Cannabis sativa21. Moreover, by taking advantage of available gene expression data26, gene expression profiling of these enzymes was performed in cannabis plant tissue at different developmental stages. The data note presented here will provide researchers with a corollary of candidate genes that will considerably accelerate future investigations on terpenoid metabolism in Cannabis sativa.
Cannabis sativa transcript sequences (n=23,630) taken from the canSat3 genome assembly (http://genome.ccbr.utoronto.ca/)21 were annotated with Blast2GO 4.0.727 using NCBI blastx and InterProScan databases. Terpene metabolism related genes were selected if found to be present in datasets downstream of the “terpene metabolic process” gene ontology category from the AmiGO 2 repository (GO:0042214), including the carotene metabolic process (GO:0016119), ent-kaurene metabolic process (GO:0033331), ent-pimara-8(14),15-diene metabolic process (GO:1901539), isoprene metabolic process (GO:0043611), miltiradiene metabolic process (GO:1901944), monoterpene metabolic process (GO:0043692), sesquarterpene metabolic process (GO:1903192), sesquiterpene metabolic process (GO:0051761), terpene biosynthetic process (GO:0046246), and terpene catabolic process (GO:0046247)28.
Gene expression profiles from cannabis plant tissue at different developmental stages were downloaded from the NCBI GEO repository (https://www.ncbi.nlm.nih.gov/geo/). Gene expression heatmaps and unsupervised hierarchical clustering were performed with GENE-E 3.0.21329.
Although the Cannabis sativa reference genome and transcriptome has been publicly released21, only a few genes have been characterized and surveyed for their molecular functions. To define the possible roles of these genes, 40,197 canSat3 transcript sequences were downloaded from the cannabis genome browser (http://genome.ccbr.utoronto.ca/), translated in silico, and scanned for evolutionarily conserved protein domains for functional annotation (Figure 1). To identify the genes presumably playing a role in terpenoid metabolism, annotated transcripts were filtered for gene ontology (GO) categories involved in terpene biosynthesis and catabolism using the AmiGO 2 reference database. A total of 288 transcripts representing 215 different genes were predicted to be involved in the metabolism of bisabolene, cadinene, carotene, copaene, ent-kaurene, farnesol, geraniol, germacrene, lycopene, limonene, myrcene, phytoene, pinene, squalene, and others (Supplementary table 1). Functional characterization of this subset confirmed an enrichment for GO categories involved in different terpene biosynthetic and catabolic processes (Figure 2).
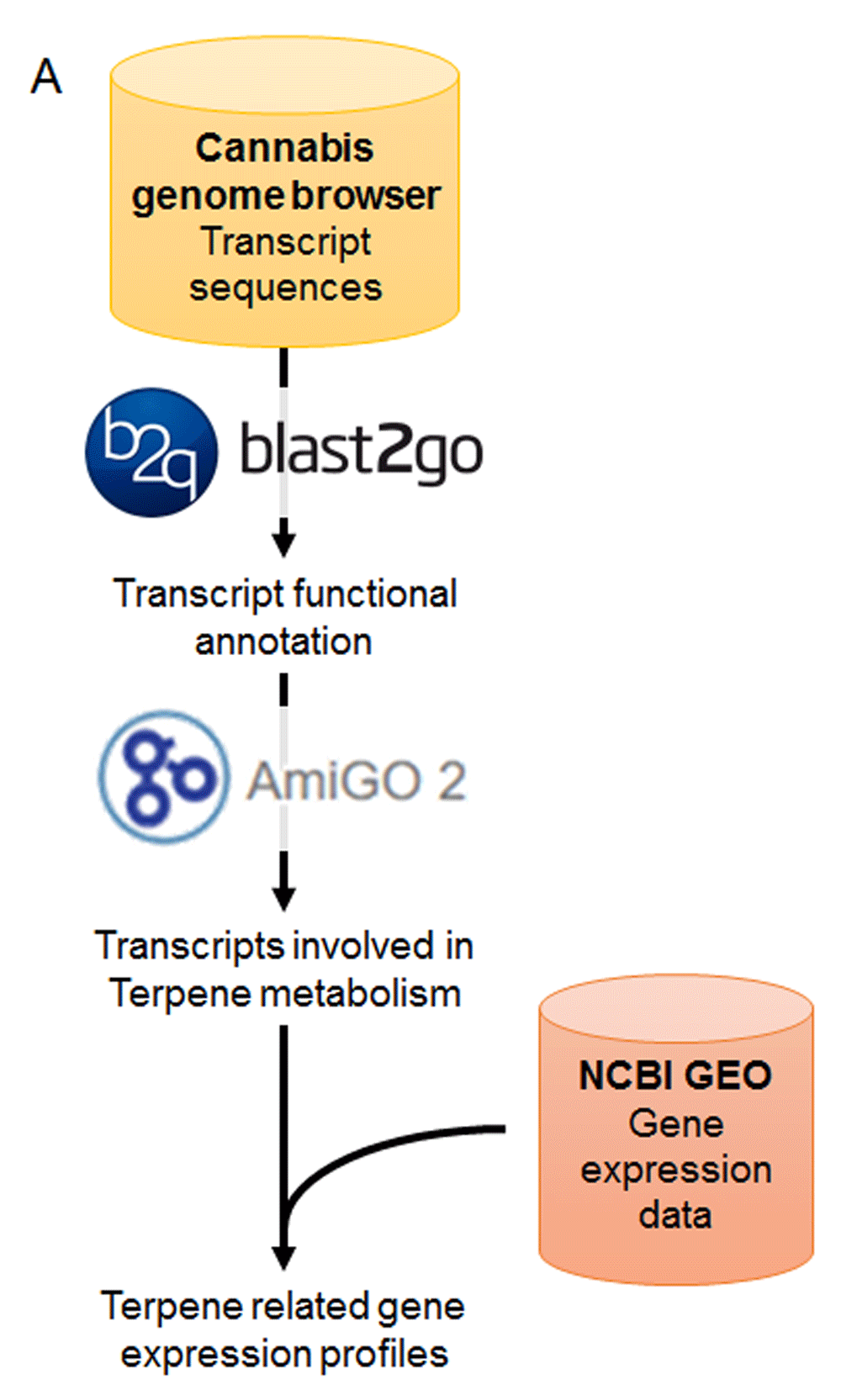
Schematic of bioinformatics pipeline utilized in this work. Cannabis sativa transcript sequences were taken from the canSat3 reference genome, functionally annotated with Blast2GO 4.0.7, filtered for terpenoid metabolism categories (AmiGO 2), and integrated with gene expression data downloaded from NCBI.

Functional enrichment analysis of putative terpenoid metabolism related transcripts taken from the canSat3 reference genome. Enrichment for terpene biosynthesis and catabolism is shown for Biological Process (A) and Molecular function (B) Gene ontology categories.
Terpenoids are produced by several plants species and in several types of plant tissue as defense against predators30. Similar to other biological compounds, their abundance directly correlates with the expression levels of the enzymes involved in their metabolism. To this end, gene expression analysis of genes likely to be involved in terpenoid metabolism was performed using previously published datasets (Figure 3; Supplementary table 2)26. Notably, unsupervised hierarchical clustering identified four gene clusters. Cluster 1 genes display high expression in roots and stems; cluster 3 genes in hemp flowers; cluster 4 genes in leaves and flowers. Cluster 2 genes were constitutively expressed in all tissues. These results highlight which enzymes are expressed by specific tissues and will provide a strong rationale for further investigations on the molecular basis of terpenoid metabolism.

Heatmap showing relative expression values (log2 RPKM) of putative terpenoid metabolism related genes from cannabis plant tissue taken at different developmental stages (shoot, root, stem, young and mature leaf, early-, mid- and mature-stage flower). Five gene clusters were defined in accordance to unsupervised hierarchical clustering.
The active principles inside plants have been exploited by humans for centuries, with Cannabis sativa being one of the oldest ever used for medicinal purposes31. Surprisingly, contrary to whole plant extracts, medicinal products containing exclusively THC have been found to lack efficacy and lead to unbearable side effects8,9. These results arise from the fact that these products lack other important co-factors typically found in the Phyto-complex, such as terpenoids and other cannabinoids10 that contribute to the synergistic effects seen with whole plant extracts.
While genes involved in cannabinoid biosynthesis have been widely investigated19–22, the gene network controlling terpenoid metabolism is only recently being elucidated, with genes encoding (-)-limonene synthase and (+)-α-pinene synthase being the only two characterized23. To this end, Cannabis sativa transcripts21 have been scanned for evolutionarily conserved protein domains and annotated according to their presumptive molecular function. As a result, 215 evolutionarily conserved genes were predicted to be involved in terpenoid metabolism. Furthermore, in silico gene expression profiling26 of these enzymes in cannabis plant tissue at different developmental stages highlighted different gene clusters with peculiar expression patterns. For instance, cluster 3 genes (Figure 3) displayed high expression specifically in hemp flowers, which could be of great interest as different cannabis strains harbor different entourage effects.
Since the current cannabis reference transcriptome is still at preliminary stages21, it is very likely that false negatives have caused important transcripts to still be missing. For example, the two genes encoding for (-)-limonene synthase and (+)-α-pinene synthase23 align on the same transcript predicted to encode for Myrcene synthase (PK25781.1 in Supplementary table 1), and therefore cannot be discriminated. Unfortunately, to overcome this issue at whole genome level we need the complete version of the reference transcriptome to be available. Until that time, researchers are forced to validate single transcripts with classic low-throughput technology, such as molecular cloning followed by Sanger sequencing.
Nevertheless, the data presented here will ease future investigations on terpenoid metabolism in Cannabis sativa by providing researchers with a collection of candidate genes. For instance, one of these genes was predicted to encode for β-bisabolene synthase (PK05069.1 in Supplementary table 1). Bisabolene is being used as an antimicrobial agent32, as well as a biofuel33. However, prior to this report nothing was known about the gene network controlling its metabolism in Cannabis sativa. As soon as future studies will integrate gene expression data with chemical analysis, the complete molecular scenario underlying terpenoid metabolism will be revealed.
Processed gene expression data can be found in the NCBI GEO repository (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE93201.
Supplementary table 1. Evolutionarily conserved terpenoid metabolism transcripts
List of putative terpenoid metabolism genes obtained with Blast2GO.
Click here to access the data.
Supplementary table 2. Terpenoid metabolism gene profiling in different tissues and developmental stages
Gene expression matrix of predicted terpenoid metabolism genes. Expression units are expressed in RPKM.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a useable and accessible format?
Partly
References
1. Wang Q, Jia M, Huh JH, Muchlinski A, et al.: Identification of a Dolabellane Type Diterpene Synthase and other Root-Expressed Diterpene Synthases in Arabidopsis.Front Plant Sci. 2016; 7: 1761 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Is the rationale for creating the dataset(s) clearly described?
Partly
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a useable and accessible format?
Yes
References
1. Booth JK, Page JE, Bohlmann J: Terpene synthases from Cannabis sativa.PLoS One. 2017; 12 (3): e0173911 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Plant metabolism
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 06 Feb 17 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)